Alan DenAdel
@alandenadel
Computational biology PhD student at @BrownUniversity. Previously at @illumina. he/him
ID: 1025373559189848064
03-08-2018 13:31:21
280 Tweet
258 Followers
1,1K Following

antisense. Omar Wagih bioRxiv bioRxiv and medRxiv are undergoing a platform upgrade that has led to unanticipated performance issues. Apologies for the inconvenience - these should be resolved soon.

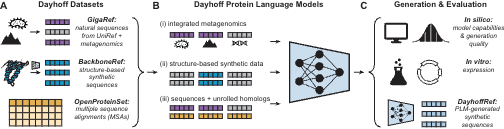
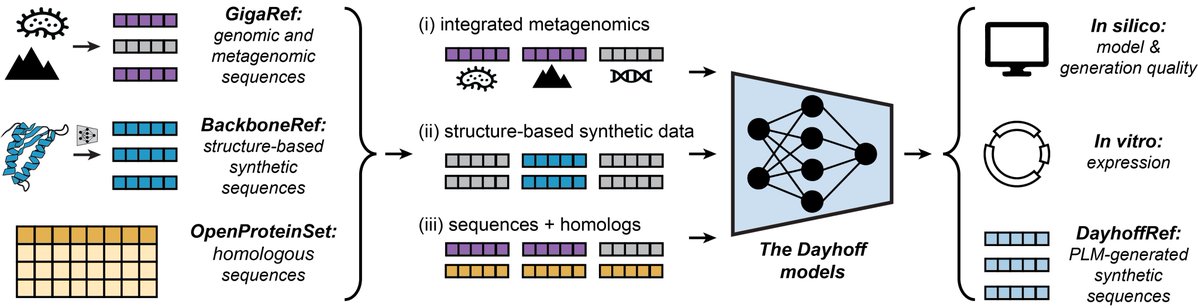
Single cell foundation models have the potential to revolutionize many areas of biology. How does the composition of training data affect the performance of these models? In my intern project Microsoft Research New England, we identify patterns that can augment next-generation models.




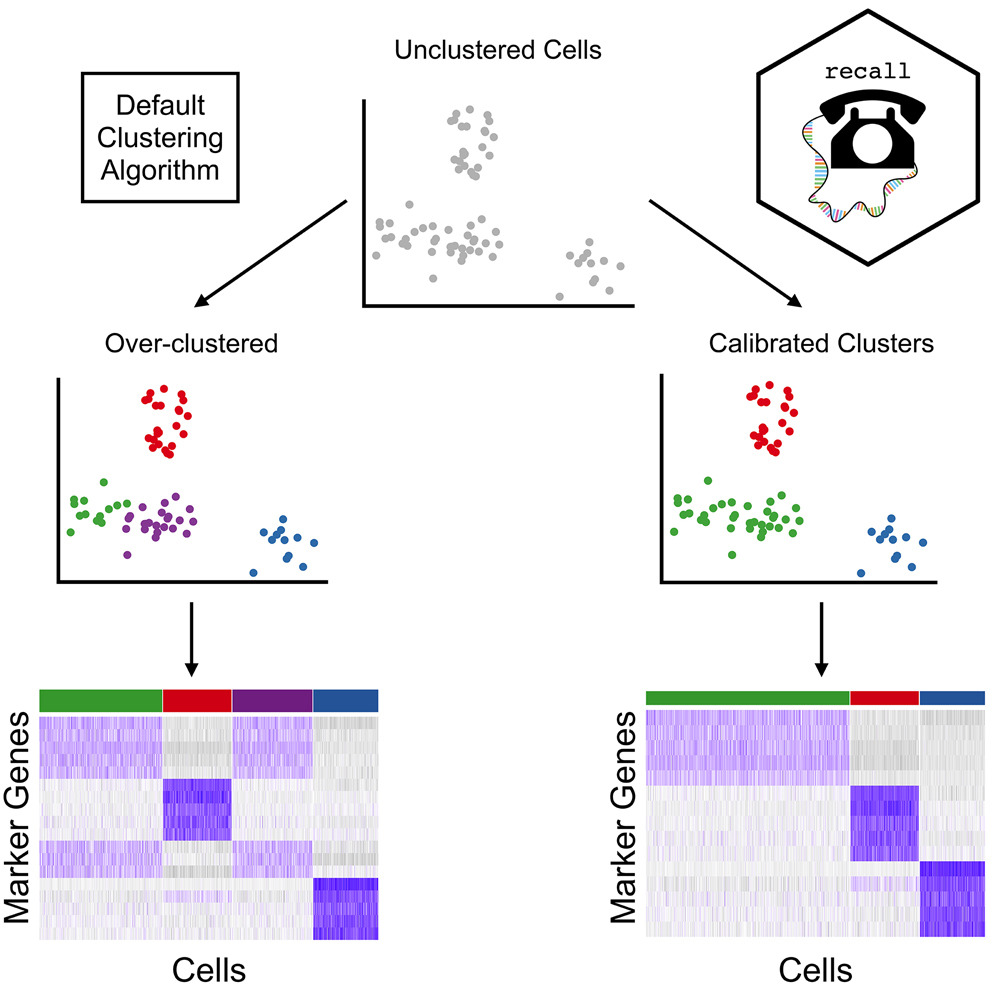
Great to see our paper presenting recall, a framework which calibrates clustering for the impact of data "double-dipping" in single-cell studies, out today in AJHG! Congratulations, Alan DenAdel and co-authors!

Over-clustering of single-cell RNA-seq data can produce spurious results. Alan DenAdel, Lorin Crawford, & co of AJHG latest study introduce recall, a new method, protecting against over-clustering & enables rapid analysis of single-cell RNA-seq data: cell.com/ajhg/abstract/…

Our paper has been published Nature Genetics! Through new statistical methods, we shed light on fundamental questions about cellular response to genetic perturbations. Our work is a substantial advance towards rigorous characterization and comparison of massive perturbation atlases.


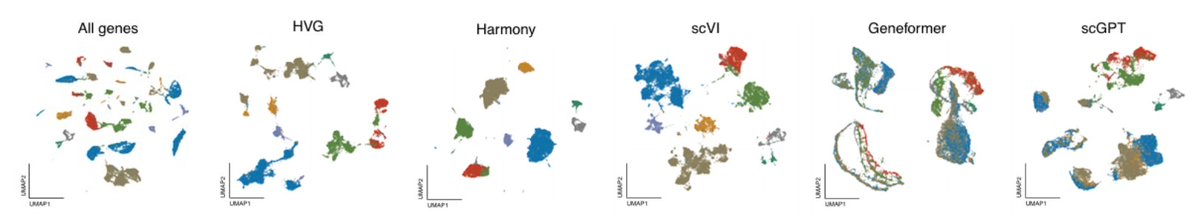
How well do single-cell foundation models perform w/o finetuning? Our work Genome Biology shows that in zero-shot settings, scGPT and Geneformer often underperform traditional methods, raising questions about their utility for biological discovery. 📰 genomebiology.biomedcentral.com/articles/10.11…

Defining “what is good” is central to both AI and Science. Despite massive efforts towards building the Virtual Cell, our recent Nature Biotechnology article reflects on whether current gold standards (i.e., evaluation metrics) still hold up. nature.com/articles/s4158…