Alexander Dilthey
@alexdilthey
Bioinformatics, genetics and population genomics. Professor @HHU_de. MHC/HLA, graphs, long reads, SARS-CoV-2, microbiome. Own opinions.
ID: 844449476
24-09-2012 22:56:17
2,2K Tweet
1,1K Followers
2,2K Following

Daria Frolova's paper in plasmid relatedness now out in MGen! She uses rearrangement distances & shared seq content to build a relatedness network of plasmids, identifies hubs which are likely transposons, and then clusters related plasmids. microbiologyresearch.org/content/journa… Microbiology Society




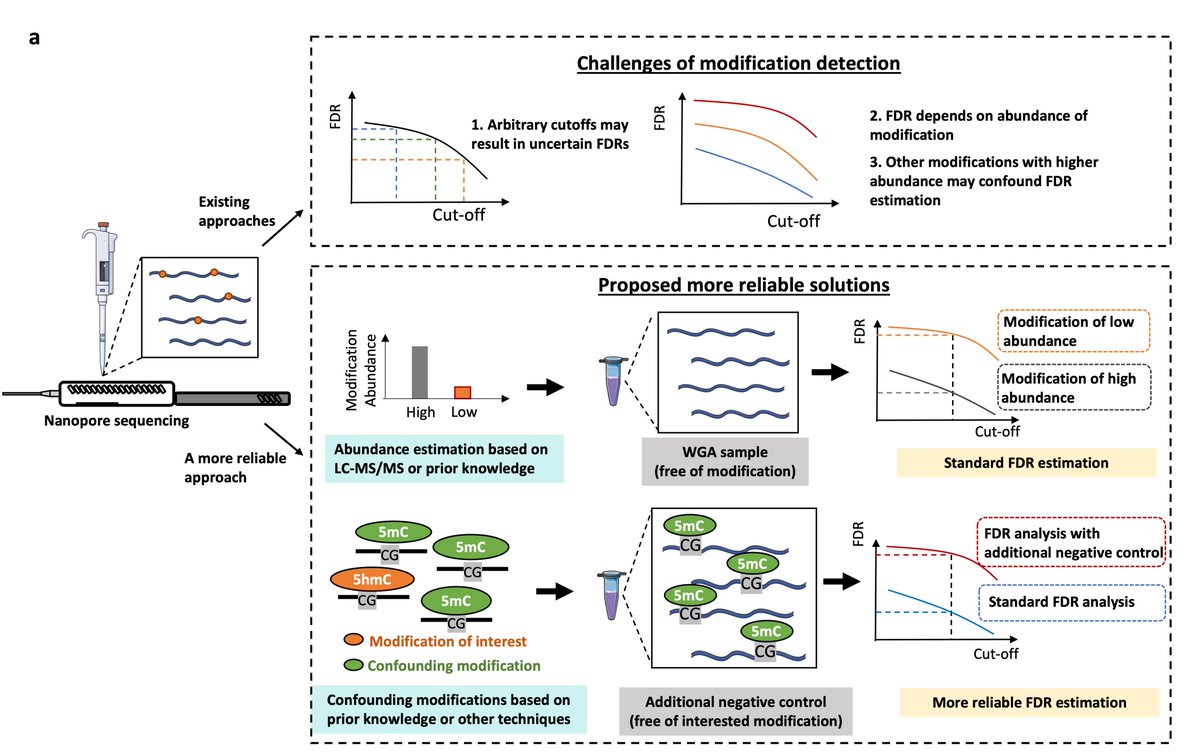
Preprint alert 🚨 Cautions in the use of Oxford Nanopore sequencing to map DNA modifications: officially reported “accuracy” ≠ reliable mapping in real applications. We performed a critical assessment of nanopore sequencing (across different versions of models) for the detection of

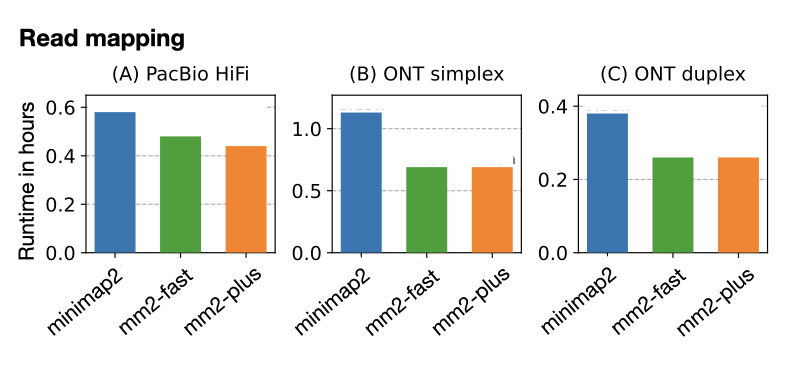
New method speeds up genome alignment of complete and near-complete genome assemblies in minimap2. Can also be useful for faster read mapping Github: github.com/at-cg/mm2-plus DOI: doi.org/10.1101/2024.1… Led by Ghanshyam Chandra, collab with Sanchit Misra (I am hiring) Mohd Vasimuddin Intel



We found that activating programmed necrosis through genetic or pharmacological interventions might be an effective anti-cancer-strategy in pancreatic cancer. Thanks to all collaborators and funders! Mathias Heikenwälder Heinrich-Heine-Universität Düsseldorf Maximilian Reichert nature.com/articles/s4146…

The 3rd annual Symposium on Big Data Algorithms for Biology (BDBio) is happening this May! Join us for insightful talks and discussions on comp-bio topics Register, submit abstract, and see program details at bdbio.in IISc Bangalore IISc Computational and Data Sciences Centre for Brain Research, IISc


A decade ago, we had thousands of bacterial genomes. Now, we have millions. How to scale computational methods? Our paper in Nature Methods answers this: use evolutionary history to guide compression and search. …From terabytes to tens of GBs… w/@Baym Zamin Iqbal et al. 🧵1/




Excited to share our paper on somatic variant calling in HLA genes and genes related to antigen presentation across a cohort of 136 patients with diffuse gliomas published in Cancer Immunology Research: doi.org/10.1158/2326-6… A thread ... #Bioinformatics #Immunology #CancerResearch



🚀 We’ve launched the Night Science Institute! Our mission: to champion a cultural shift in science toward embracing the creative scientific process – as a vital complement to rigorous hypothesis testing. Watch the launch video with Itai Yanai: drive.google.com/file/d/1qpuxad…