Alexander Hanzl
@alexanderhanzl
SNSF postdoctoral fellow @LabThoma at @FMIscience
PhD with @Georg_e_Winter at @CeMM_News
Chemical biology, phenotypic screening, structural biology.
ID: 1272804176641363969
16-06-2020 08:12:50
69 Tweet
199 Followers
252 Following

In their new Cell Reports paper, researchers from former CeMM PI & MedUni Wien Assoc. Prof. Joanna Loizou (Joanna Loizou) demonstrate how inhibiting the POLΘ enzyme can stop the growth of #cancer cells with mutations in #BRCA1 ✂️🧬 ➡️ bit.ly/3tBIc2l



E3-Specific Degrader Discovery by Dynamic Tracing of Substrate Receptor Abundance CeMM @georg_e_winter Alexander Hanzl Fischer Lab #E3Specific #Degrader #Tracing #Receptor #Abundance pubs.acs.org/doi/10.1021/ja…

Great paper by the group of @georg_e_winter in J. Am. Chem. Soc.. They establish a luciferase-based screening platform that allows the systematic identification of new E3 ligases amenable to molecular glue design as well as their degradation targets. doi.org/10.1021/jacs.2…


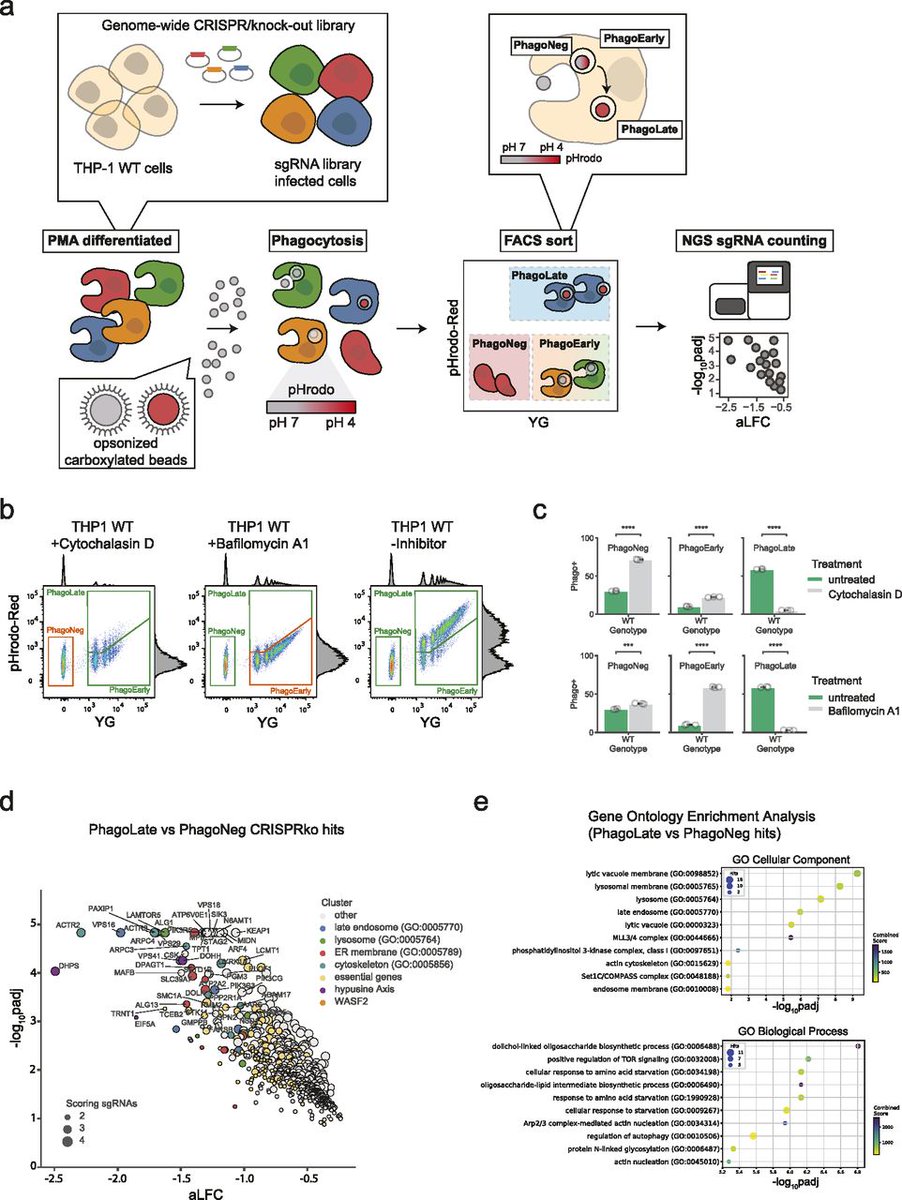
A paper from CeMM Superti-Furga Lab Patrick Essletzbichler Giulio Superti-Furga uses genome-wide FACS based reporter screening to generate a broad view of the molecular network regulating phagocytosis in human cells. life-science-alliance.org/content/6/4/e2…

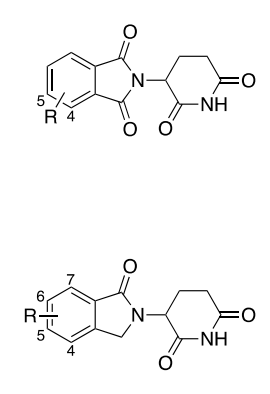
Thrilled for our Orpinolide story to see the light of day! Huge thanks to the @georg_e_winter lab, Waldmann lab, Luca Laraia and everyone that contributed Hana Imrichova (Novotna), @FabianFrommelt, Laura Depta, Andrea, Chrysanthi Kagiou, Thomas, Cris Mayor-Ruiz, Superti-Furga Lab and Sonja Sievers! 🧵👇


Delighted to share our spotlight on a beautiful paper by Alessio Ciulli and @georg_e_winter. Alex Hanzl and colleagues combined structural and biochemical techniques with deep mutational scanning to identify residues crucial for degrader activity. nature.com/articles/s4158…

If you like #genetics in human cells you may enjoy our latest paper by Gianluca Talevi and collaborators. A ‘fishing expedition’ using logic as our guide by Gian-Luca McLelland to study triglyceride synthesis. After some plot twists it yields DIESL! nature.com/articles/s4158…

Today in @bioRxiv, we describe Click Editors (CEs) which combine DNA-dependent DNA polymerases, HUH endonucleases and RNA-programmable nickases for versatile genome editing without double-strand breaks 🧬. A study co-led with Connor Tou. A 🧵 (1/14): biorxiv.org/content/10.110…

Thank you very much to the Austrian Life Science Association ÖGMBT. It is a great honor to receive this award and wouldn't have been possible without the whole @georg_e_winter lab and our collaborators Alessio Ciulli Fischer Lab

Excited to share 2 preprints, together showing how mechanisms of a molecular glue & E3 ligase cancer mutations serendipitously converge to cause neomorphic protein degradation in distinct contexts. Highlights below: 1/17 #1: biorxiv.org/content/10.110… #2: biorxiv.org/content/10.110…


🔬 Targeted Protein Degradation: A new adapter molecule expands the therapeutic potential around the cell's waste disposal system, CeMM PI @georg_e_winter's team published a new study in Nature Communications : go.nature.com/3zn9Ia2 Read more ➡️ bit.ly/4cmwYn6


Excited to share our latest preprint. Lead by Sevi (Sevi Durdu) we asked how transcription factors (TFs) with short and abundant motifs drive specific cell fates. Our test cases: Two bHLH factors that bind to seemingly similar E-boxes. 1/7 biorxiv.org/content/10.110…

How do past viral infections influence future viral diseases? We found that SARS-CoV-2 recovery protected from severe influenza A virus disease. But how come? There seemed to be some antigen-independent immunological memory going on.. Paper Immunity: authors.elsevier.com/c/1js3h3qNrUxu…


In back to back preprints from Arc Institute, the labs of LukeGilbert and Patrick Hsu present two distinct platforms (COMET and COMBINE) for systematically exploring combinations of protein domains, each of which yielded a diverse range of CRISPR-guided transcriptional effectors