Amber Tang
@amberzqt
Research Scientist @instadeepai /Deep Learning for Bio/ genomic LLMs/
ID: 3420132208
13-08-2015 12:53:56
14 Tweet
125 Followers
79 Following

Be on the lookout around campus today for a troupe of Alex Ganns on the loose this Halloween Cold Spring Harbor Laboratory @wsbsbot


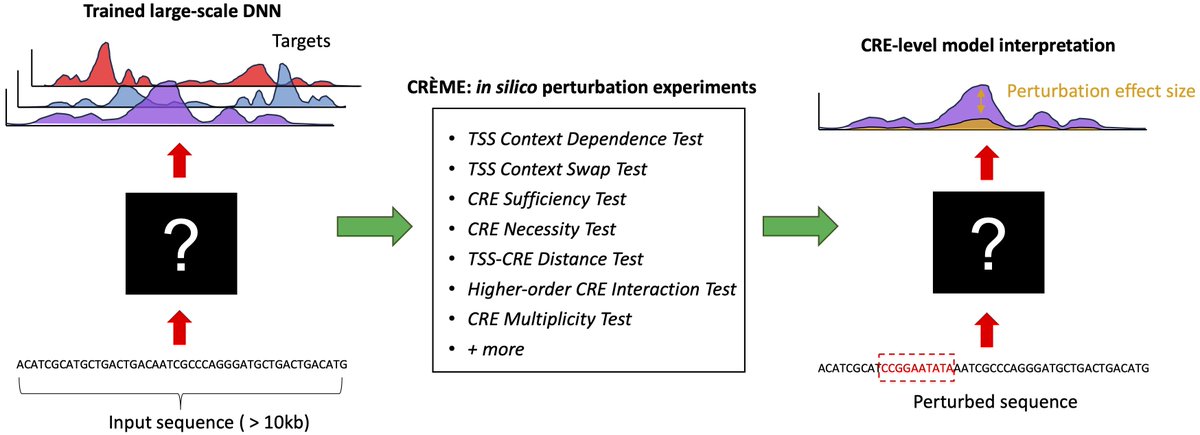
New work led by Shushan Toneyan and Ziqi (Amber) Tang (Amber Tang) on evaluating deep learning models for predicting epigenomic profiles. #RegulatoryGenomics 1/5 biorxiv.org/content/10.110…





Excited to share new work on "Interpreting cis-regulatory mechanisms from genomic deep neural networks using surrogate models” led by Evan Seitz, jointly advised by me and Justin B. Kinney and in collab with David M. McCandlish Paper: biorxiv.org/content/10.110… Docs: squid-nn.readthedocs.io


Excited for #MLCB2023! Check out talk by ShToneyan on uncovering higher-order CRE interactions from large-scale DNNs and 2 posters: revisiting inits by Chandana Rajesh and Amber Tang and new attribution method using domain-inspired surrogate models by Evan Seitz! biorxiv.org/content/10.110…

Do current genomic language models (pre-trained on whole genomes) learn a foundational understanding of biology in the non-coding region of human genomes? A new evaluation led by Amber Tang suggests not yet! 1/N paper: biorxiv.org/content/10.110…



Excited to sponsor and participate in the Machine Learning for Computational Biology Conference #MLCB2024, kicking off today in Seattle! Watch Thomas Pierrot explain how InstaDeep is advancing Generative AI for Genomics! 🧬 📽️ Live here: bit.ly/47hkcVb




Thrilled to open-source the dataset behind our Nature Machine Intelligence cover paper! 🧬 The ChatNT training data is now open-source on Hugging Face. It's the first large-scale dataset for training conversational agents on biological sequences. A thread on what's inside 👇

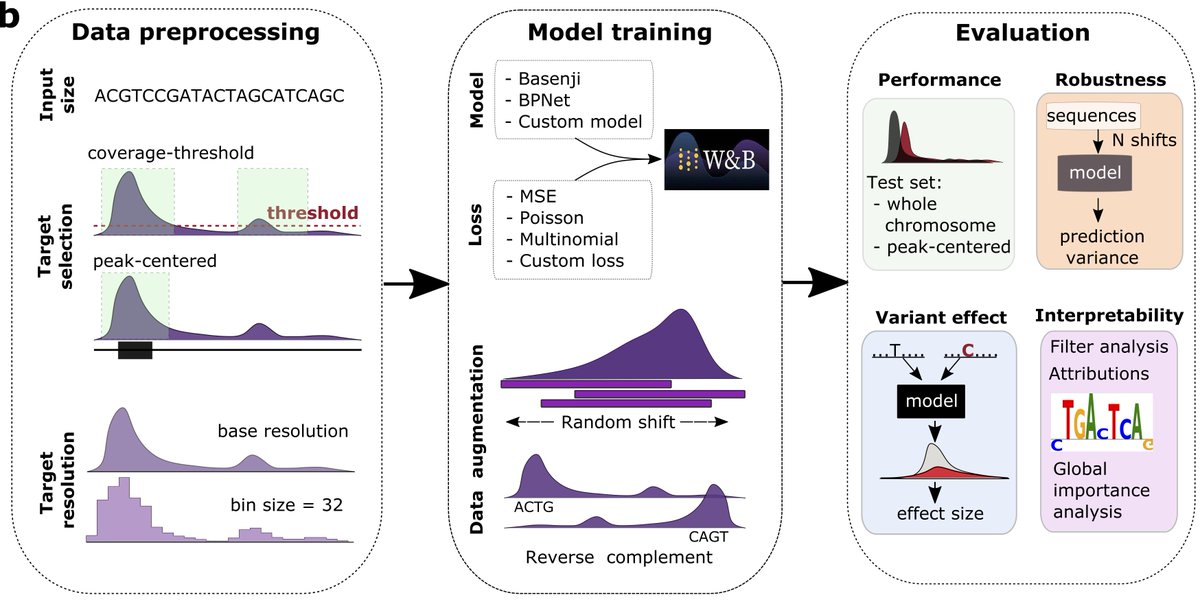
Our work on "Evaluating the representational power of pre-trained DNA language models for regulatory genomics" led by Amber Tang with help from Nirali Somia & Steven Yu is finally published in Genome Biology! Check it out! genomebiology.biomedcentral.com/articles/10.11…