Haoyu Zhang
@andrewhaoyu
Investigator @NCIEpiTraining, Statistics, Genetics and Genomics, Data Science. Views are all mine
ID: 754114452430073856
https://andrewhaoyu.github.io/ 16-07-2016 00:44:31
370 Tweet
555 Followers
271 Following


Excited to share our preprint, describing a method for heritability partitioning with GWAS sumstats that significantly improves upon S-LDSC Led by the fantastic Hui Li, and co-supervised by Xihong Lin #ASHG24 poster 4089F medrxiv.org/content/10.110…



Current polygenic risk scores require binning of ancestry groups but we know ancestry is a continuum. Our study led by Yunfeng Ruan🌻 #StandWithUkraine🌻 introduces DiscoDivas, a new generalizable PRS framework across the ancestry spectrum medrxiv.org/content/10.110… medRxiv PRIMED Consortium



Our new PRS paper is out! We introduce a method to create regularized ensemble PRS on GWAS sumstats. Individual-level data is no longer needed for sophisticated ensemble learning Zijie Zhao Stephen Dorn Paper📰:biorxiv.org/content/10.110… Software🧑💻:github.com/qlu-lab/PUMAS


Excited to share our latest work in Cell Genomics! Using a novel MR method integrated with AlphaFold3, we identify Alzheimer’s-associated proteins with 3D structural changes. cell.com/cell-genomics/… Andrea Baccarelli Gary W Miller ColumbiaPublicHealth Cell Genomics

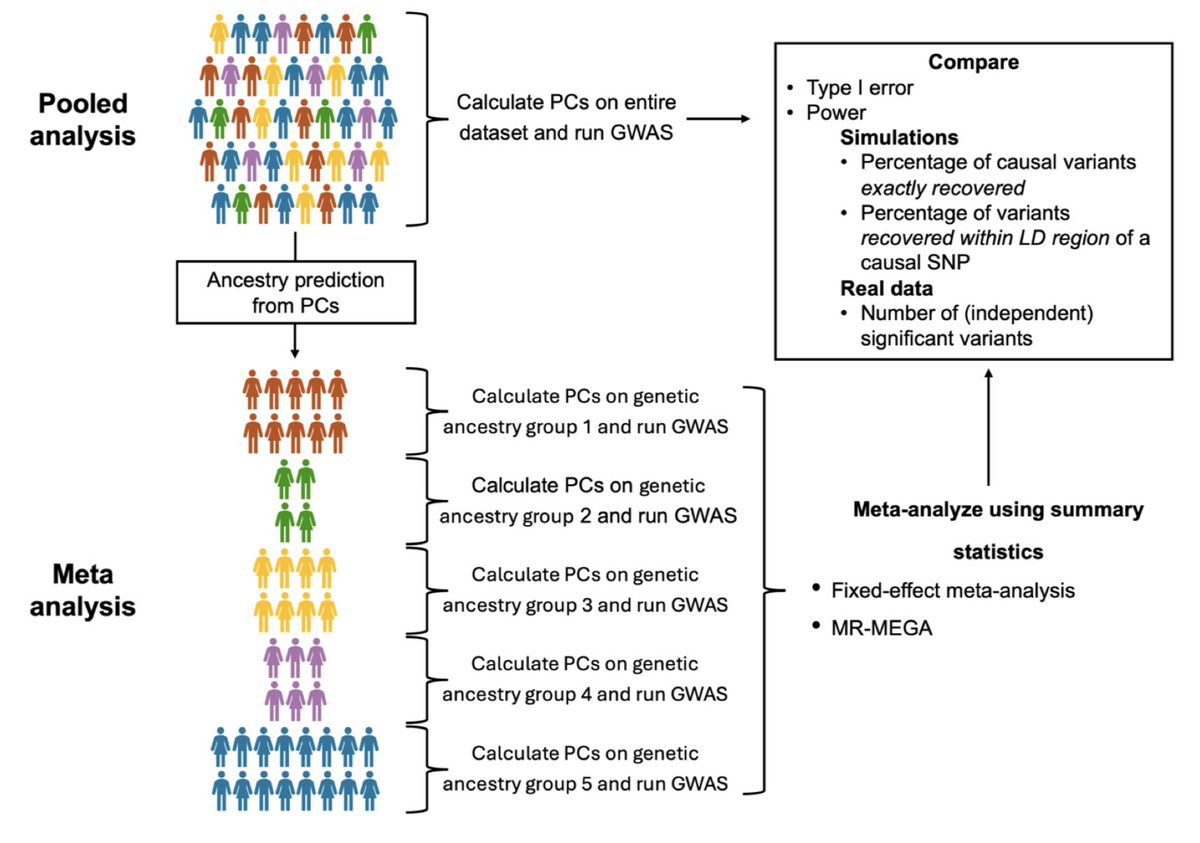
Excited to share our new manuscript on evaluating multi-ancestry GWAS methods using large-scale simulations & real data from UK Biobank (N≈324K) & All of Us (N≈207K)! My first thesis paper under the guidance of Pete Kraft & Haoyu Zhang. Available at: medrxiv.org/content/10.110…

Very excited to share our preprint led by Michael Levin Satoshi Koyama Jakob Woerner & with Scott Damrauer assessing genome-wide pleiotropy of >1,000 clinical traits across ~1.7M individuals with nearly 30K locus-trait associations! medrxiv.org/content/10.110… medRxiv [1/3]
![Pradeep Natarajan (@pnatarajanmd) on Twitter photo Very excited to share our preprint led by <a href="/MGLevin/">Michael Levin</a> <a href="/skoyamamd/">Satoshi Koyama</a> <a href="/jakob_woerner/">Jakob Woerner</a> & with <a href="/damrauer/">Scott Damrauer</a> assessing genome-wide pleiotropy of >1,000 clinical traits across ~1.7M individuals with nearly 30K locus-trait associations!
medrxiv.org/content/10.110… <a href="/medrxivpreprint/">medRxiv</a>
[1/3] Very excited to share our preprint led by <a href="/MGLevin/">Michael Levin</a> <a href="/skoyamamd/">Satoshi Koyama</a> <a href="/jakob_woerner/">Jakob Woerner</a> & with <a href="/damrauer/">Scott Damrauer</a> assessing genome-wide pleiotropy of >1,000 clinical traits across ~1.7M individuals with nearly 30K locus-trait associations!
medrxiv.org/content/10.110… <a href="/medrxivpreprint/">medRxiv</a>
[1/3]](https://pbs.twimg.com/media/GpN2beJWMAAyF55.jpg)

Delighted to collaborate with J. Hu and Hongyu Zhao to show that genetic mechanisms leading to a high CAD polygenic risk score are heterogeneous, which may have implications for prevention journals.plos.org/ploscompbiol/a… PLOS Comp Biol