
Anna Nordin
@annanordin96
PhD student in the Cantù lab studying Wnt signaling and chromatin structure during development and disease
ID: 1542509105213911046
30-06-2022 14:04:04
36 Tweet
149 Followers
160 Following

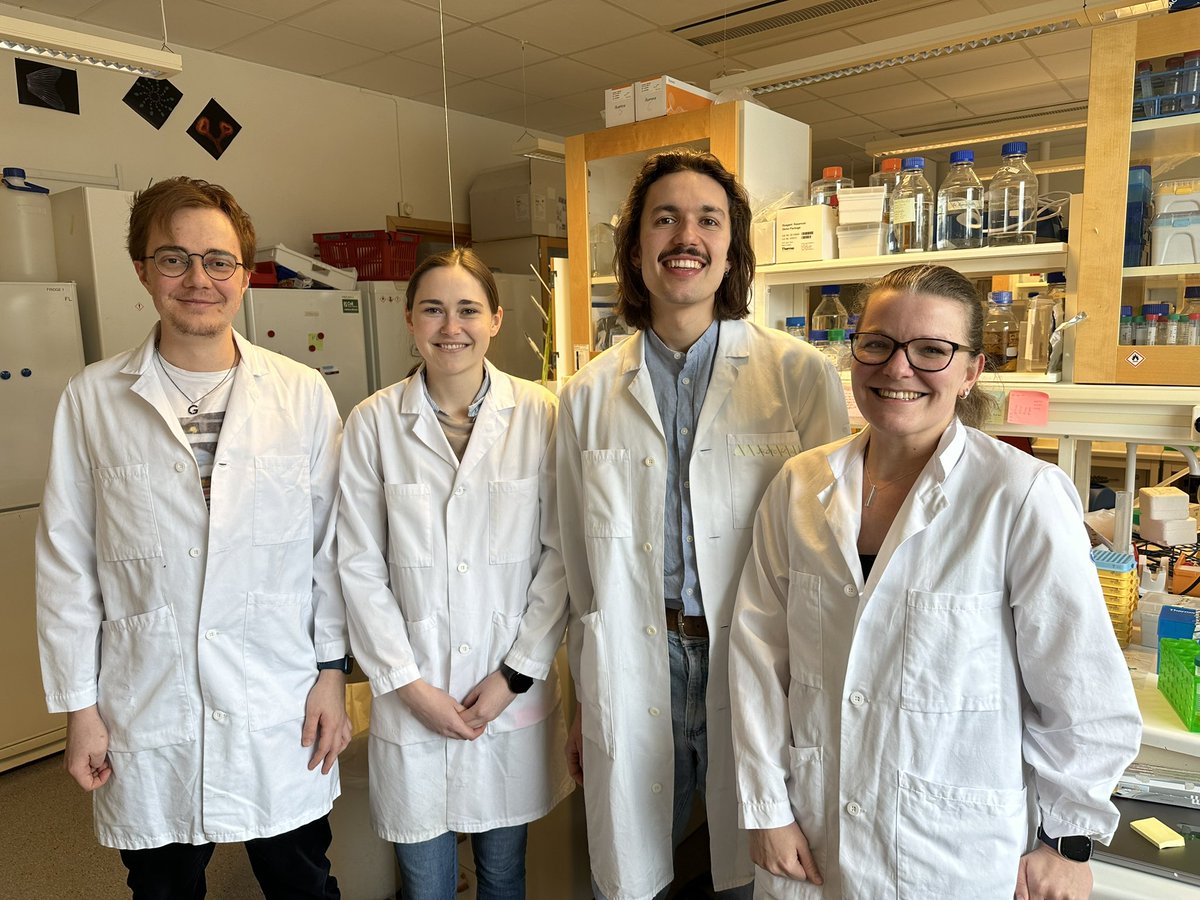
Anna Nordin and I from Claudio Cantù lab are thrilled to have hosted Kim Boonekamp and Johannes Werner from Boutroslab to learn CUT&RUN LoV-U and start an exciting collaboration! For details 👇 doi.org/10.1242/dev.20… Wallenberg Centre For Molecular Medicine_Linköping Linköpings universitet


The #UNCOUPLING of #WNT target gene expression previously on bioRxiv is now with #PeerReview in Experimental Cell Research By Simon Söderholm Amaia Pierfrancesco Pagella Valeria Ghezzi Gianluca Zambanini Anna Nordin CantùLab is Wallenberg Centre For Molecular Medicine_Linköping sciencedirect.com/science/articl…

Deepest dive into the data I've ever taken and wow did we find some unexpected things! This was such an exciting project to be a part of, as always it was amazing to work with Pierfrancesco Pagella, Gianluca Zambanini, and Claudio Cantù at Linköpings universitet Wallenberg Centre For Molecular Medicine_Linköping

The Time-Resolved Genomic Impact of #WNT/β-catenin signaling Cell Systems Cell Press cell.com/cell-systems/f… We show -β-catenin repositions over TIME ⏰ -How this impacts global transcription⬆️⬇️ -Preceding and following chromatin dynamics🧬 Efforts driven by Pierfrancesco Pagella


Ever looked at your ATAC-seq signal and thought: there must be a better way to model this than peaks or windows? Happy to present ChromatinHD, a method that models scATAC+RNA data using the raw fragments. With @BartDeplancke Olga Pushkarev biorxiv.org/content/10.110…

Super happy that my first first-author paper of my PhD is now out in Genome Biology! Try filtering out the suspect list peaks to improve your CUT&RUN analysis - and we are always grateful for feedback! Thanks to the amazing Gianluca Zambanini, Pierfrancesco Pagella and Claudio Cantù.








I'm so excited to have this out! Please use the data, and we love feedback! A huge team effort as always, especially thanks to Gianluca Zambanini (You can blame me for the ❤️🧡💛💚💙💜🩷 though 😅) Claudio Cantù Pierfrancesco Pagella


Excited to share our latest preprint on benchmarking peak-calling methods for CUT&RUN biorxiv.org/content/10.110… | Amin Noorani In this work, we compared four popular methods—MACS2, SEACR, GoPeaks, and LanceOtron, in a subset of 4DNucleome samples profiled for 3 histone marks.




