
Dr. Anna Ramisch
@annaramisch
CompBio Postdoc interested in HumanGenetics, NeuroEpigenomics & Addiction @unige_en
ID: 2924903080
16-12-2014 12:29:31
366 Tweet
274 Followers
429 Following

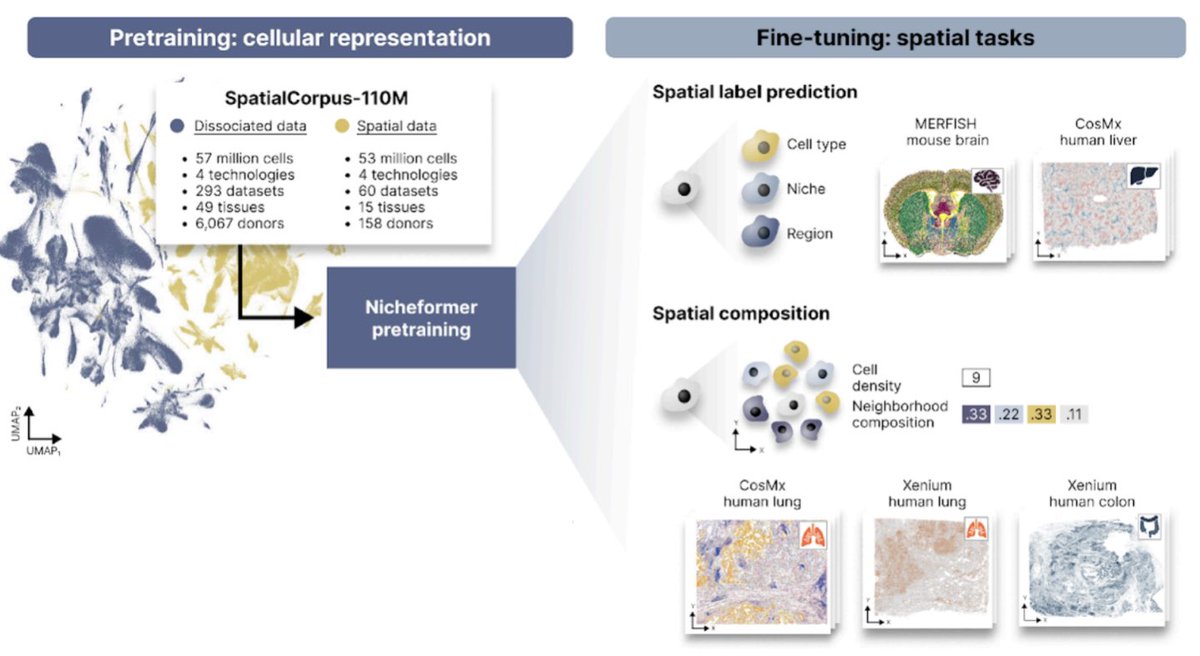
Excited to share Nicheformer! Led by Alejandro Tejada Lapuerta & @AnnaCSchaar, Nicheformer is a foundation model for single-cell & spatial omics. Its innovation is going beyond disassociated analysis to capture & predict local tissue context at single-cell level. biorxiv.org/content/10.110…

Inspired by Lior Pachter and team's exploration of scRNA-seq pipelines, we face similar challenges in scATAC-seq, only magnified! After 5 years of tackling these inconsistencies with my advisor @JunhyongKim , Here is a summary (and some solutions😃) 1/n #Genomics #Bioinformatics

Excited to share with the single-cell community the most recent work of Tomàs Montserrat Ayuso and AnnaEsteveCodina from CNAG, something we think will be a game-changer in #scRNAseq quality control: biorxiv.org/content/10.110… bioRxiv Get ready for the tweetorial! 1/10

Ana Ana Viñuela is motivating us to look at eQTLs beyond gene counts to transcript counts from long read sequencing data Ana et al used Oxford Nanopore to generate 🧬 data #BoG24


Characterizing >100,000 sequences in primary neurons and organoids and deep learning to decode regulatory activity. Amazing work from Chengyu Deng, Sean Whalen, Katie Pollard, Nowakowski_Lab and others as part of #PsychENCODE in Science Magazine. science.org/doi/10.1126/sc…

How do enhancers work over distances that sometimes exceed megabases? Excited to share our work led by Grace Bower where we uncover a unique sequence signature globally associated with long-range enhancer-promoter interactions in developing limb buds: biorxiv.org/content/10.110… 1/



Figeno, my visualization tool for genomics🎨🧬, is now published in Bioinformatics! doi.org/10.1093/bioinf… It can generate publication-quality figures for sequencing data along genomic coordinates: bigwig, HiC, Oxford Nanopore data with base modifications, and WGS with CNAs and SVs.


Des scientifiques de l’#UNIGE ont identifié les séquences génétiques régulant l’activité des gènes responsables de la croissance osseuse. Guillaume Andrey Fabrice Darbellay #science #médecine #recherche #génétique ow.ly/cfek50SizmR


‼️New Epigenetics Review ‼️ Read here at Genes & Development where we discuss brief history of various epigenetic modulator classes 🧬, their links to multiple disease types 😷, and their potential for (combined or novel) therapeutic targeting 💊! m.genesdev.cshlp.org/content/early/…


We are looking for a bioinformatician Collège de France CIRB_CdF in Paris to analyse genomic/epigenetic datasets🧬 in the context of a synergy European Research Council (ERC) long term program. See below and apply😎 RT🙏


I am excited to share our new preprint with Ana Viñuela, Dr. Anna Ramisch, Andrew Brown, and Manolis Dermitzakis! We used direct RNA long-read seq on 60 LCLs from the 1000 Genomes Project to study genetic regulation of transcript abundance & RNA modifications (m6A). researchsquare.com/article/rs-461….


Really happy for Aline Réal to be getting this out. Major implication for me is I've always assumed gwas hits will be explained by expression as it has to mediate then. But here Aline Réal shows these expression effects can take many forms, not all picked up by common tech


New benchmarking study of 8 pipelines and 5 widely used methods for scATAC data analysis..includes embedding, graph and partition-based metrics and useful recommendations by data complexity and downstream tasks. Snakemake workflow: github.com/RoseYuan/sc_ch… genomebiology.biomedcentral.com/articles/10.11…

I'm very excited to share that I’ve started as a group leader Karolinska Institutet. I am looking for curious, enthusiastic and collaborative postdocs and assistants (pre-PhD) to join us to decipher regulation of hypoxia cell states that govern systems oxygen homeostasis 1/3

Delineating the effective use of self-supervised learning in single-cell genomics Nature Machine Intelligence Fabian Theis nature.com/articles/s4225…

Very excited to share that our manuscript on cis-eQTL analysis of adipose tissue gene expression in 2,344 samples is now out in Nature Genetics rdcu.be/d5ov4 See thread from lead author Sarah Brotman below
