Anton Bushuiev
@antonbushuiev
PhD student working on machine learning for molecule discovery at CTU Prague
ID: 1557303807931043841
10-08-2022 09:52:28
31 Tweet
68 Followers
297 Following

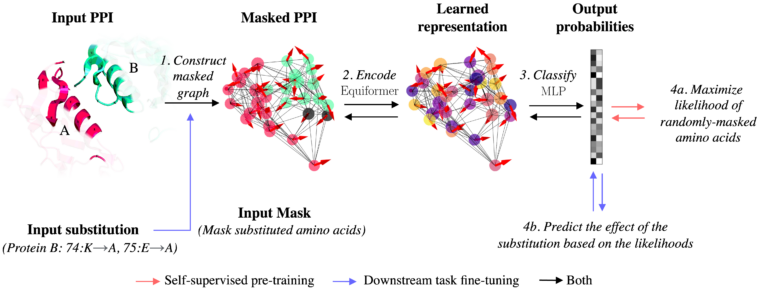
Scientists CIIRC ČVUT Loschmidt Laboratories, Masaryk University IOCB Prague and ICRC have developed a new method to improve the computational design of biotherapeutics. Their new machine-learning method enables more efficient design of proteins with better interaction properties. bit.ly/3UUdHmO


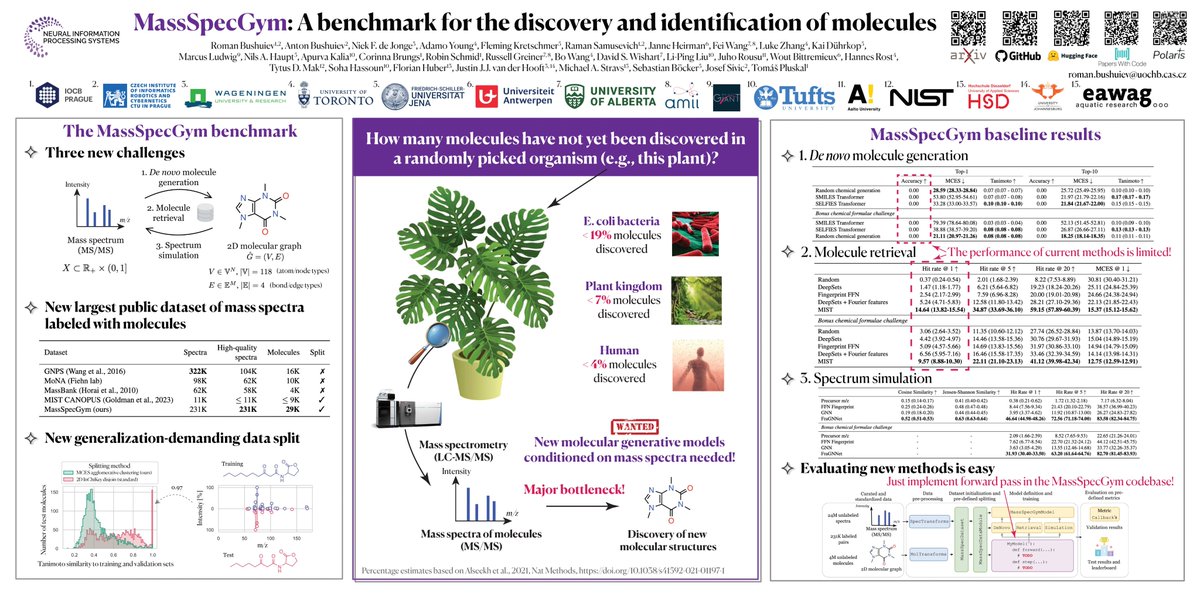
Can machine learning help unlock the vast potential of unexplored molecular structures in biological and environmental samples through mass spectrometry data analysis? IOCB Prague "MassSpecGym: A benchmark for the discovery and identification of molecules" • With less than





Working on predicting molecular structure from mass spec data? Check out MassSpecGym from Roman Bushuiev, Anton Bushuiev, and the team! This might be one of the most well-documented datasets on Polaris right now. MassSpecGym has the largest publicly available collection of 231K




The MassSpecGym team presenting the poster at #NeurIPS2024! Fei Wang Adamo Young Roman Bushuiev Anton Bushuiev Raman Samusevich



🤝 In April 2024, brothers Roman and Anton Bushuiev from the teams of Tomáš Pluskal IOCB Prague and Josef Šivic CIIRC ČVUT initiated a collaboration among 14 research institutes across the globe to benchmark #AI methods for the discovery of molecules from mass spectrometry data.


Back in July 2023 we organized a small bioML symposium at IOCB Prague, and it turned out to be a very pleasant and successful event. This summer we are following up with a great line-up of speakers. Please register for free, deadline May 30. tinyurl.com/bioMLPrague25



⚛️ Tomáš Pluskal from IOCB Prague, together with his student Roman Bushuiev and colleagues from #CIIRC CTU, Josef Šivic and Anton Bushuiev, have developed a machine learning model called #DreaMS – which accelerates the analysis of previously unknown molecules.