Archit Singh
@architex_97
Doctoral researcher working on problems in Statistical Genetics with @EleZeggini at @MunichDS @HelmholtzMunich and @TU_Muenchen
ID: 1404163364213854208
13-06-2021 19:47:33
40 Tweet
70 Followers
457 Following

At this time a fascinating paper from Danny Reinberg lab demonstrated differential histone dilution kinetics in active versus repressed chromatin during replication tinyurl.com/5n8vp3tn. The authors did not connect this to diffusion, but to me the signs were clear (4/8)

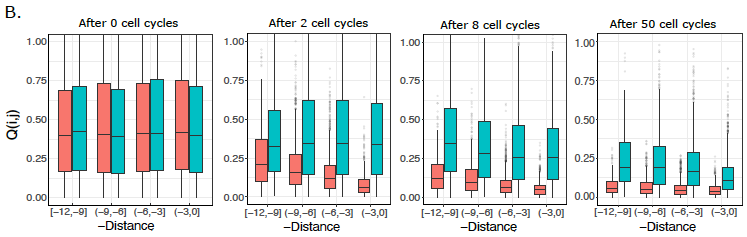
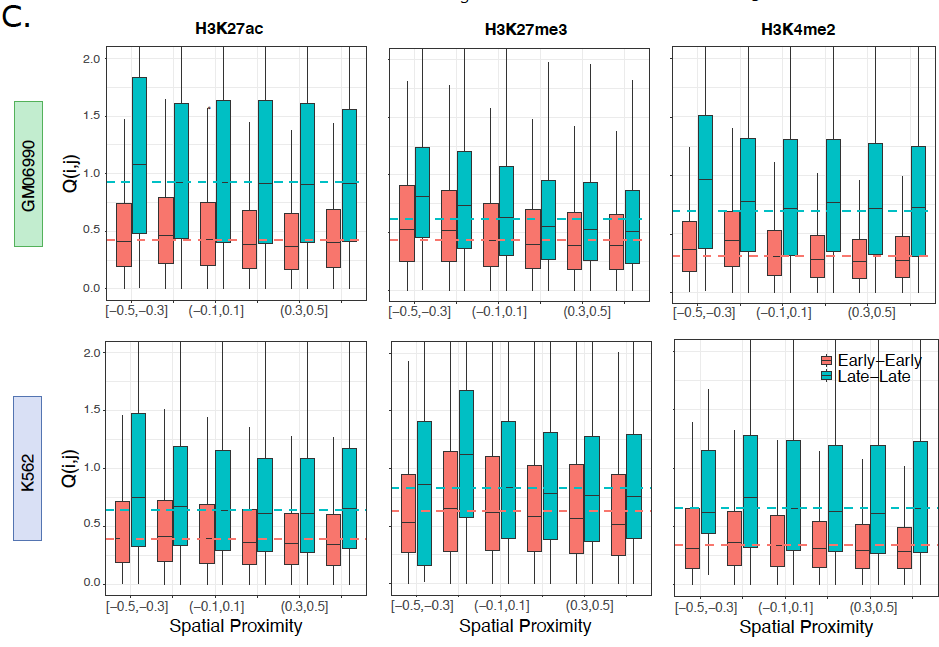
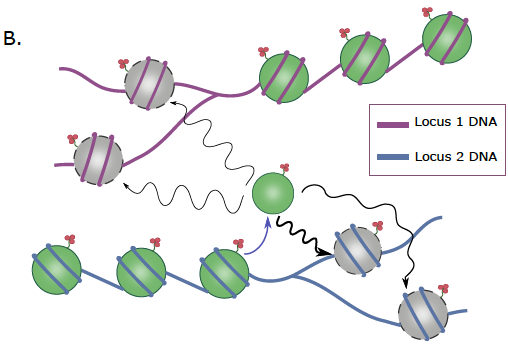
Together with Archit Singh, a fantastic student in my lab, we developed a lattice model for the replication fork with diffusive dynamics of histones. Incorporating larger diffusivity in active chromatin quantitatively recapitulated the previous experimental observations (5/8)
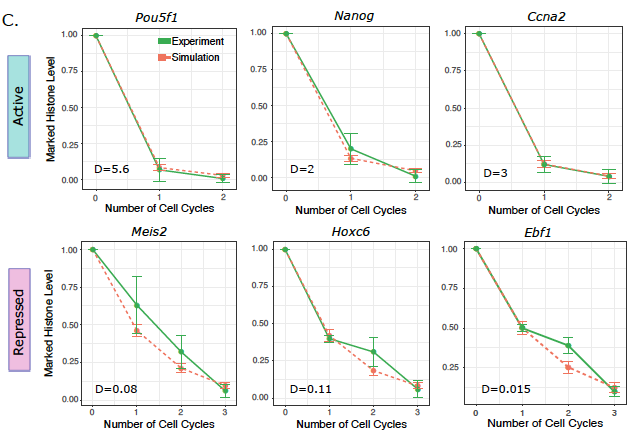








Very happy that our new work from Living Machines-NCBS National Centre for Biological Sciences on histone diffusion, challenging the conventional idea of 100% accurate histone-mark inheritance during replication, is now out in PLOS Comp Biol ! tinyurl.com/5hbnkaj2

Can we combine inhibitors of the #circadian clock and lineage correlations to detect presence of cell cycle gating by the clock? Excited about our new manuscript from National Centre for Biological Sciences, Living Machines-NCBS showing how this might be possible! A short thread (1/6) tinyurl.com/46pub9rh







Translated to German 🇩🇪 Prakhar Gupta India in Germany German Embassy India