Ashleigh Lister
@ashleighslister
Senior Research assistant @EarlhamInst in Tech Dev focusing on Single Cell and spatial transcriptomics protocols for use in plant tissues in @EI_single_cell
ID: 1154052678
06-02-2013 13:45:57
194 Tweet
233 Followers
278 Following

Come visit me at my poster Monogram Network to learn more about our collaboration with Earlham Institute Ashleigh Lister on spatial transcriptomics in wheat 🌾
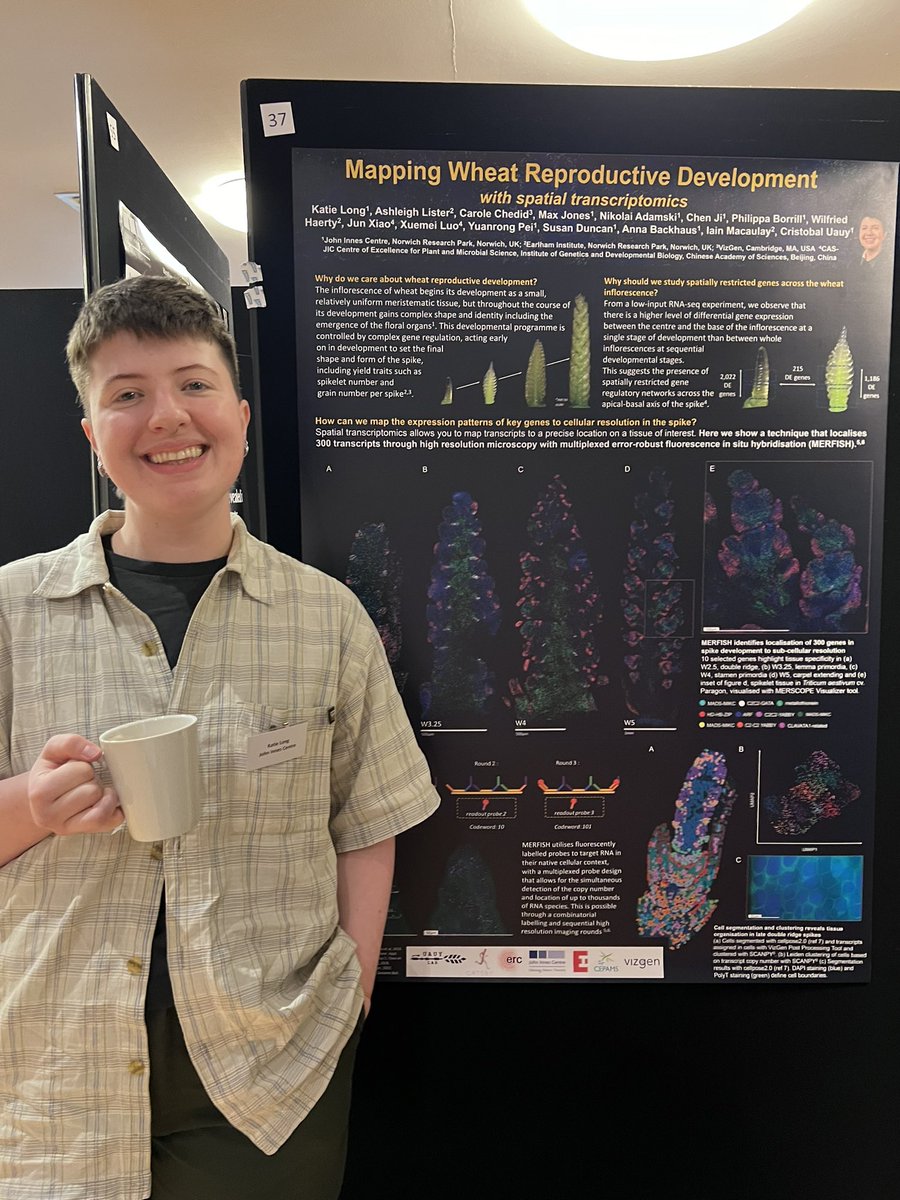

Monogram Network @SophJCarpenter Fabulous work Sophie! Spatial transcriptomics data is the result of fantastic team work from Katie Long and Ashleigh Lister. Check out Katie's poster #37. #monogram24





Super excited to be welcoming delegates to the first Earlham Institute Vizgen Spatial Transcriptomics in plants workshop. It's going to be a great couple of days! With Ashleigh Lister and Carole


Looking forward to hearing lots of great science talks at the #EISingleCell24 symposium here Earlham Institute over the next couple of days. It will be great to show participants around our labs and meet others interested in single cell and spatial transcriptomics technologies.

It's the second day of #EISingleCell24 symposium with @biouea, and we're looking forward to lots more #singlecell discussions, including: 🌱 #SpatialTranscriptomics in wheat 🔎 Expression Atlas resource 💧 Open-source microfluidics 🧬 #Longread sequencing in health and disease


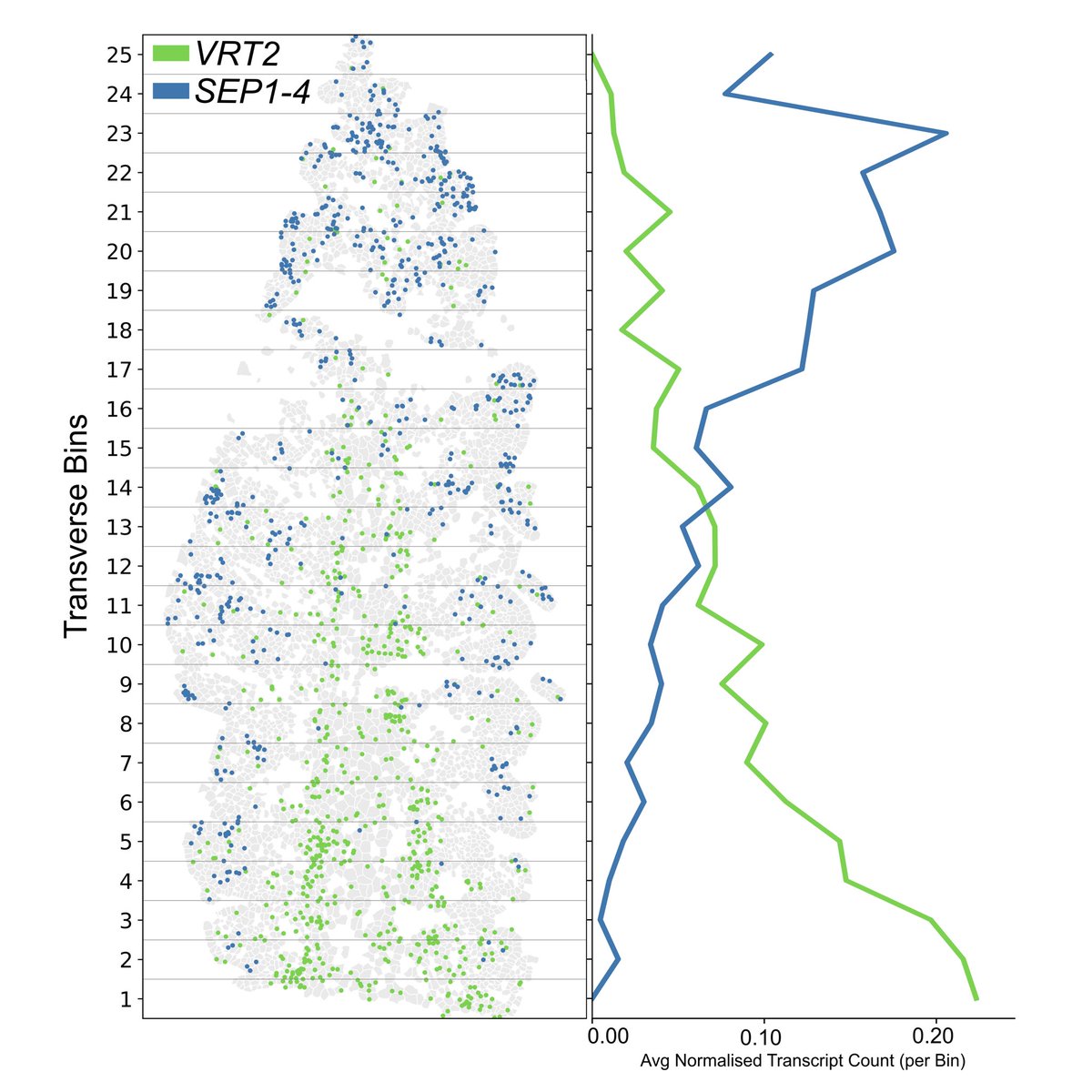
#EISingleCell24 Up next we're looking forward to hearing Katie Long from @johninnescentre, discuss her work with Ashleigh Lister at the Earlham Institute, using #SpatialTranscriptomics in wheat spikelet development. Find out more here 👇 okt.to/bnJpl1

Welcoming delegates to Earlham Institute Single Cell RNAseq Laboratory course with a bit of theory. Great to have such a diverse group of backgrounds from attendees.


💻Join Vizgen & Global Engage #Webinar 🗓️15 Nov | Friday | 09:00 AM GMT Experts from Earlham Institute will share their journey working with the MERSCOPE & offer tips for optimizing the platform for tricky samples. 🔗hubs.ly/Q02RWZj80 👉Register to view on-demand!




Read our new paper on spatial transcriptomics in wheat spikes! Earlham Institute EI Single Cell now @whatchamacaulay.bsky.social Incredible analysis by Katie Long John Innes Centre and a lovely collaboration. Great to have a proof of our capability in this new technology in plant tissues. biorxiv.org/content/10.110…

1/9 I am excited to share our pre-print on spatial transcriptomics in wheat spikes in collaboration with Ashleigh Lister. Using MERFISH, we resolved the expression of 200 genes to cellular resolution. Check out the pre-print 👉tinyurl.com/c7t3kncf



A great thread from Katie Long at John Innes Centre discussing their collaboration with Ashleigh Lister and the Earlham Institute's #singlecell team, using our MERFISH platform to spatially profile 200 wheat spike genes. #SpatialTranscriptomics Vizgen Take a look 👇