
Žiga Avsec
@avsecz
Research scientist at @GoogleDeepMind.
Previously PhD student @gagneurlab working on deep learning methods for genomics.
ID: 4631868921
28-12-2015 11:49:20
402 Tweet
1,1K Followers
514 Following

We're hiring a research scientist to join our Quantum Chemistry and Materials team ⚛️🚨 The team is working on using machine learning to better our understanding of the universe, down at the level of quantum physics. See: deepmind.com/blog/simulatin… Share: boards.greenhouse.io/deepmind/jobs/…

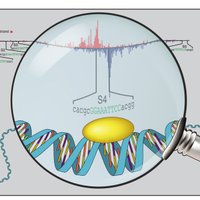
In our latest work spearheaded by Khyati Dalal Khyati Dalal , we use interpretable deep learning of genomic sequence information to tackle a long-standing question: How do signaling pathways regulate transcription in a cell-type-specific fashion? (biorxiv.org/cgi/content/sh…) (1/8)

I had an incredible time chatting with Žiga Avsec, a brilliant mind at Google DeepMind, for our Superbio.ai Scientist Spotlight series. blog.superbio.ai/superbio-scien…

📢 MIA Spring ’24 Speaker Series ⏰ March 13th Wednesday, 9AM ET 💬 Žiga Avsec (Žiga Avsec) and Jun Cheng (@jun90cheng) from Google Deepmind 💡Primer and seminar on: Alpha Missense: proteome-wide variant effect prediction (deepmind.google/discover/blog/…)


Announcing AlphaFold 3: our state-of-the-art AI model for predicting the structure and interactions of all life’s molecules. 🧬 Here’s how we built it with Isomorphic Labs and what it means for biology. 🧵 dpmd.ai/3URDiNo





Solving International Maths Olympiad (IMO) level problems has been a grand challenge for AI systems. Happy to share that a new solver (AlphaProof) developed by our team Google DeepMind and our geometry solver (AlphaGeometry) were able to solve 4 out the 6 IMO 2024 problems!

🧵 1/9 Delighted to announce that #DeepRVAT is out today in Nature Genetics! Here's a recap of our tweetorial from the preprint, with updates for some exciting new results. It's been a joy to work with Eva Holtkamp, as well as gagneurlab and Oliver Stegle.
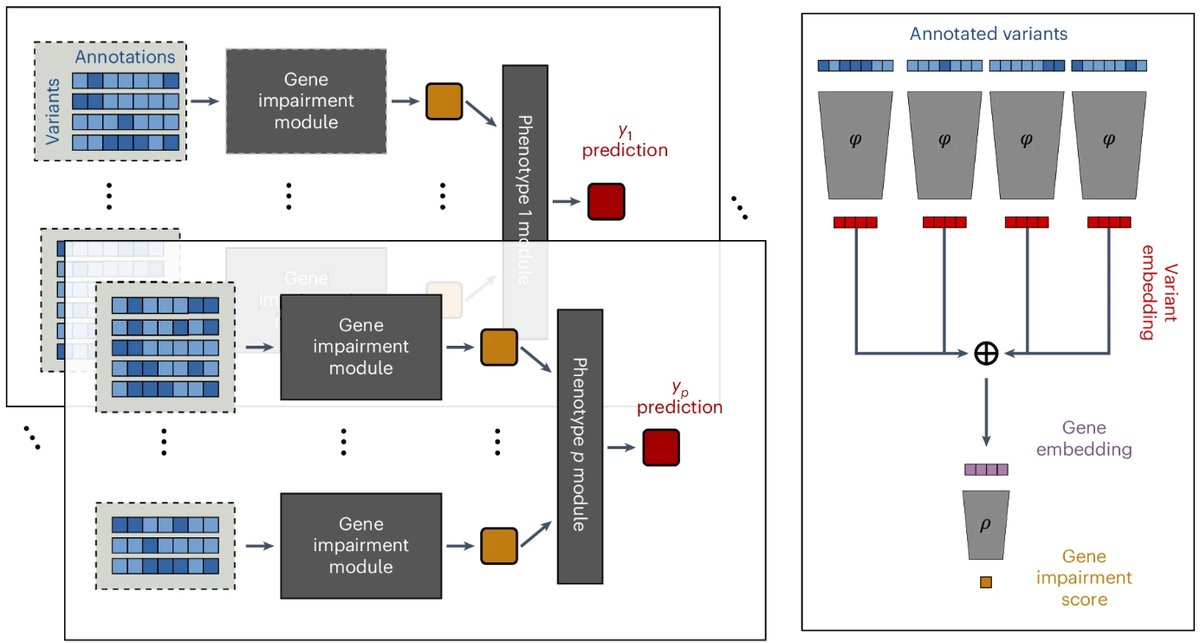

Really pleased to see DeepRVAT published. A new method for rare variant genetics by Eva Holtkamp & Brian Clarke in collab with gagneurlab. I like to think of DeepRVAT as one network on top of another, harnessing variant annotations from bags of models- AlphaMissense and alike.


If AlphaFold was proof of AI’s potential, it is now also prelude – to a future of incredible progress, driven by AI, shaped by human ingenuity, and guided by a collective sense of responsibility and safety. Massive congrats to Demis Hassabis and John on winning the Nobel Prize!


I’m excited to co-host the new #AIxBio conference alongside Žiga Avsec, Noelia Ferruz, Patrick Hsu, Ben Lehner, mary jane, Debora Marks, Tingying Peng, @CarolineUhlerat at Wellcome Genome Campus/ Wellcome Sanger Institute! We’ll focus on modellingz and designing DNA, RNA, proteins, and cells, as well as




