
Azam Hussain
@azamhussai70792
#MachineLearning applications for protein modeling. PhD candidate @UMich in the @charleslbrooks lab
ID: 1674797242278969344
https://www.linkedin.com/in/azamh/ 30-06-2023 15:09:28
252 Tweet
95 Followers
184 Following

The first episode of The Baker Lab Podcast is live. Meet Mohamad Abedi, who went from being a Palestinian refugee to a postdoctoral researcher. His journey proves that science can be a creative and transformative process. Apple: podcasts.apple.com/us/podcast/the… Spotify: open.spotify.com/show/32pWWpKzO…
















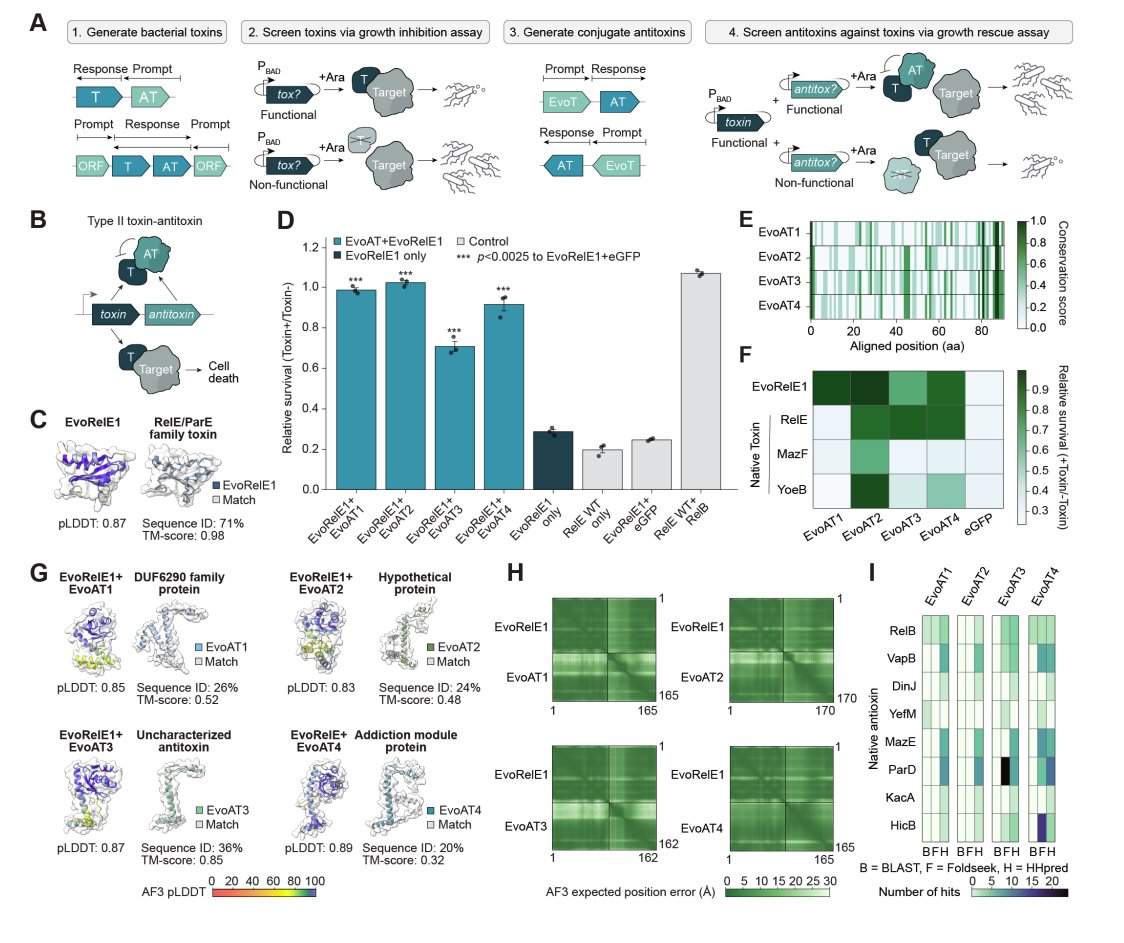
A protein's position in the genome in relation to other genes is informative about function, so a genomic language model can be prompted to generate - toxin/antitoxin pairs - anti-CRISPR proteins + a database of 120B synthetic base pairs Aditi Merchant Samuel King Eric Nguyen Brian Hie


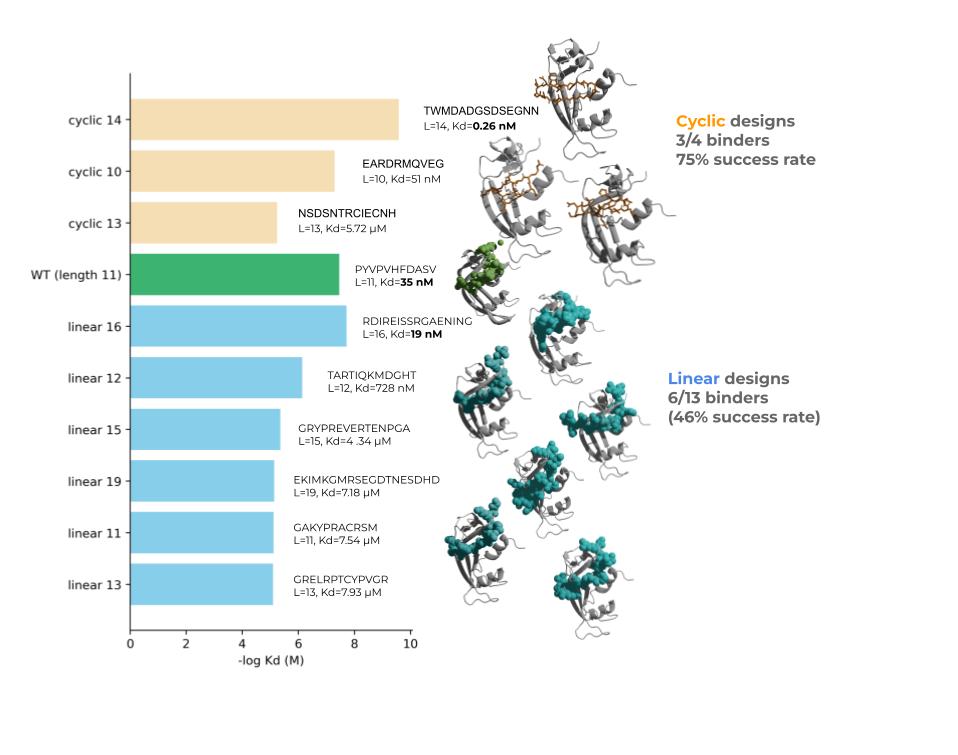


![fajie yuan (@duguyuan) on Twitter photo 🚀 Announcing ColabSaprot v2!
🧬 Train your own protein language models instantly - no ML or coding expertise required.
Everyone can do it in a few minutes!
📺 Video Tutorial: [youtube.com/watch?v=nmLtjl…]
💻 Try it: [colab.research.google.com/github/westlak…]
📄Paper: [biorxiv.org/content/10.110…] 🚀 Announcing ColabSaprot v2!
🧬 Train your own protein language models instantly - no ML or coding expertise required.
Everyone can do it in a few minutes!
📺 Video Tutorial: [youtube.com/watch?v=nmLtjl…]
💻 Try it: [colab.research.google.com/github/westlak…]
📄Paper: [biorxiv.org/content/10.110…]](https://pbs.twimg.com/media/Gd9l6N4XYAAhgbO.jpg)