Bintu Lab
@bintulab
Single-cell chromatin and gene regulation dynamics for better cell engineering @ Stanford BioE
ID: 1065111872549007361
https://www.bintulab.com/ 21-11-2018 05:17:14
99 Tweet
1,1K Followers
162 Following

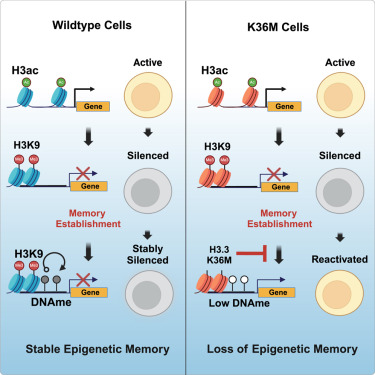
It's a double feature on epigenetic memory today! Check another amazing new preprint on single-cell chromatin compaction and memory establishment from wizard postdoc Taihei Fujimori!




Our first RNA-centric preprint is online! Congratulations to Abby Thurm and the whole team - we're so excited to continue learning about effector domains in RBPs, and to hear your thoughts about what could come next!


(1/16) Check out our Simon Gaudin paper tiny.cc/icuazz from the Canzio lab out in Science Magazine today on how tuning local cohesin trajectories enables differential readout of the clustered Protocadherin (Pcdh) locus across different types of neurons.






Lastly, I would also like to give a shout out to Nate Hathaway’s lab UNC Pharmacy for writing a beautiful preview article elegantly describing our work. Please check it out here! : shorturl.at/hycOp (4/4)