
Bob Schiffrin
@bobschiffrin
Postdoc at Astbury Centre, University of Leeds, UK.
Biochemistry, molecular dynamics, structural biology, scripting
ID: 1238055691493027840
12-03-2020 10:54:20
139 Tweet
123 Followers
324 Following
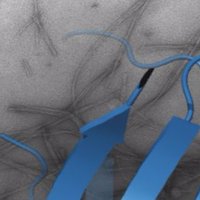



Check out our latest preprint with yiying yang Robin Corey @robincorey.bsky.social @JulienMarcoux2 Phill Stansfeld. YifL/LptM promotes oxidative maturation of the lipopolysaccharide translocon by substrate binding mimicry. biorxiv.org/content/10.110…

Anyone for PIMs? Modelling mycobacterial membranes to tackle TB: pnas.org/doi/10.1073/pn…. Chelsea Brown's first 1st author paper of her PhD, with Robin Corey @robincorey.bsky.social, @DrLizF, CHARMM-GUI, and excellently co-coordinated by matthieu Chavent.




In our just accepted paper in Angewandte Chemie with RadfordLab & Neil Ranson in The Astbury Centre we used In-Cell PELDOR/DEER complemented by CryoEM to show the BAM complex adopts a lateral closed conformation in E coli cells in presence of antibiotic darobactin-B: onlinelibrary.wiley.com/doi/10.1002/an…


Excited to see @milkowilko's structure of human arctic Aβ1-42 ex vivo from AppNL-G-F mouse brain. Check out the paper in Nature Communications rdcu.be/dcnfb to see tomograms showing fibrils in unfixed brain tissue from Conny Leistner and @drrenefrank The Astbury Centre @astbury_BSL


Finally, we can share fantastic structures of 'utilisomes' - glycan processing machines from the OM of our gut microbiota. Out today in nature rdcu.be/dd0dy Joshua White Augustinas Silale Dave Bolam & van den Berg. The Astbury Centre @Astbury_BSL @ScienceLeeds University of Newcastle




Some NMR animations to help teaching this semester: karamanoslab.com/nmr-animations. Good excuse to play with JavaScript #NMRchat

Are you interested in chaperones? Do you have expertise in biophysics or protein structure analysis (e.g. by MS, NMR, FRET)? We have a Biotechnology and Biological Sciences Research funded 3 yr postdoc position available, which is an exciting collaboration with RadfordLab and Zhuravleva Lab BioNMR@Leeds (1/3)


Just in time for Christmas! Excited to see our fibril structures published, showing changing polymorphism of IAPP during the course of amyloid assembly. As always, brilliant team effort from the amyloid team, thanks all The Astbury Centre Neil Ranson RadfordLab sciencedirect.com/science/articl…

Happy to finally share our work demonstrating the de novo design of b-barrel nanopores with complete control of the pore size and shape. Fantastic collaboration between my group VIB_StructuralBiologyBrussels, Institute for Protein Design, RadfordLab and Hiller Lab! biorxiv.org/content/10.110…


.Bob Schiffrin and I have written a preview for Molecular Cell on David Balchin's study of chaperone coordination during co-translational protein folding. Check out our preview: authors.elsevier.com/a/1jPbE3vVUPRm…, and the exciting paper: sciencedirect.com/science/articl…


Very excited to share our paper on outer membrane assembly which includes the first cryoEM structures of BAM-SurA complexes. A great team effort with Jim Horne Joel Crossley Anton Calabrese RadfordLab Neil Ranson nature.com/articles/s4146…


