Brent Chick
@brentchick
@UCSDbiosciences PhD student in the @HargreavesLab at @SalkInstitute • Interested in cell fate decisions, epigenetic gene regulation, and qBio
ID: 1448147180
22-05-2013 06:36:36
247 Tweet
407 Followers
2,2K Following


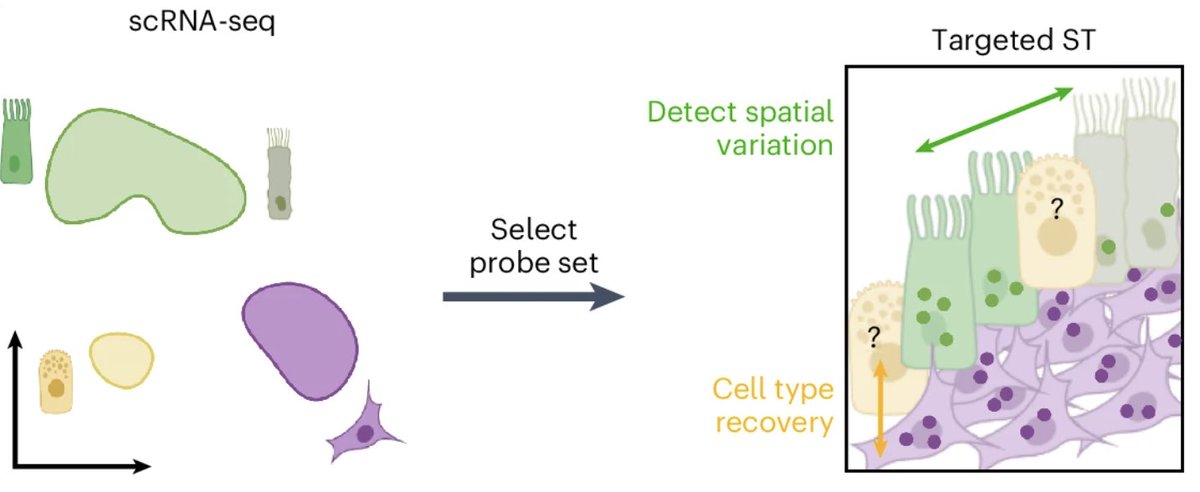
Need to design your next Xenium/CosMx/Merfish experiment? Spapros, now out Nature Methods, allows for probe set selection for target spatial tx. Led by Louis Kuemmerle, it optimizes both cell type ID & within-cell type variation. nature.com/articles/s4159… github.com/theislab/spapr…


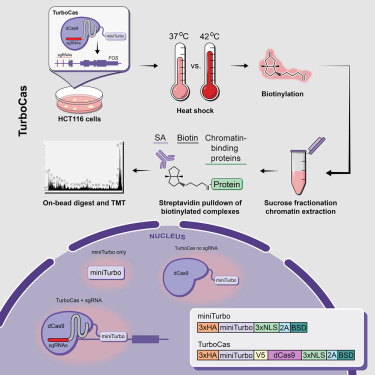
Gene regulation involves thousands of proteins that bind DNA, yet comprehensively mapping these is challenging. Our paper in Nature Genetics describes ChIP-DIP, a method for genome-wide mapping of hundreds of DNA-protein interactions in a single experiment. nature.com/articles/s4158…




Ever wondered how transcription choreographs histone modifications? Our work reveals the basis of co-transcriptional H3K36me3 by SETD2. We visualize how a histone writer coordinates with the transcription machinery! This is the magnus opus of Jon Markert tinyurl.com/setd2



Our ChromBPNet preprint out! Huge congrats to Anusri Pampari! This was quite a slog for both of us but we r very proud of this one! It is a long read but worth it IMHO. Methods r in the supp. materials. Link in the next tweet. Bluetorial coming soon .. 1/


An #OpenAccess review on current status of KRAS inhibitors, resistance drivers & potential combinations from Der Lab at UNC Channing Der Bjoern Papke & colleagues DKFZ Charité - Universitätsmedizin Berlin in Trends in Cancer As always very informative illustrations: A: Frequency of KRAS Allelic alterations across



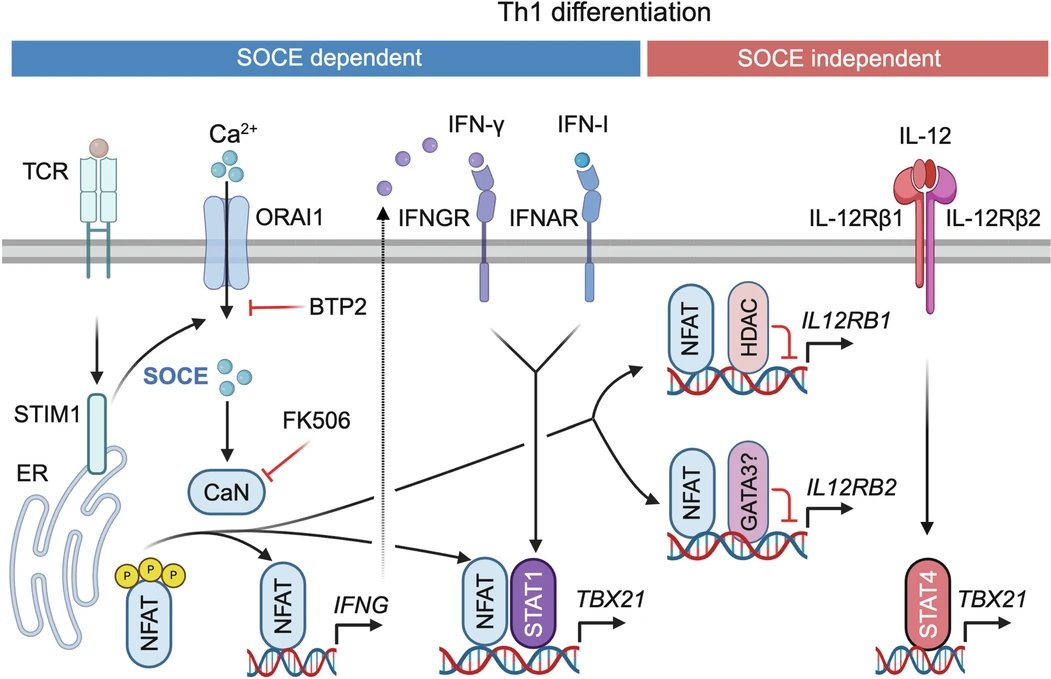
STIM1-mediated NFAT signaling synergizes with STAT1 to control T-bet expression and TH1 differentiation nature.com/articles/s4159… Nature Immunology Feske lab at NYU


Excited to share our pre-print on the molecular architecture of heterochromatin in human cells🧬🔬 w/ Jan Philipp Kreysing, Johannes Betz,Turoňová lab, Marina Lusic, Hummer lab, Beck Laboratory, Max Planck Institute of Biophysics 🔗 Preprint here: doi.org/10.1101/2025.0…