Solomon Charles
@cusolomon
Bioinformatician interested in gene regulatory network.
ID: 2185083305
17-11-2013 20:40:45
79 Tweet
95 Followers
360 Following

Attention everyone "Save the Date" and register early for our 2023 BHF Research Accelerator Symposia - Wednesday 11th October @ College Court, Leicester British Heart Foundation University of Leicester


#EMBOpopgen is back for its sixth edition, in person 13 – 19 March 2023 in Procida, Italy! Registration deadline 3rd Jan 2023 - RT please! Enza Colonna Andrea Manica Flora Jay Kelley Harris AnnaSapfo Malaspinas Benjamin Peter Leo Speidel Lucy van Dorp meetings.embo.org/event/23-pop-g… #EMBOpopgen


My lab will open its doors this summer in the Department of MCDB (MCDB Michigan). I have openings at all scales (PhD, Postdoc, Tech). Please contact me via email if you’re interested. I could not imagine a better environment to charter new frontiers in cis-regulatory biology!

The paper I am sharing today is a thoughtful philosophical perspective from Silvia Domcke & Jay Shendure proposing a new organizational framework for single cell data, as an alternative to e.g Human Cell Atlas Compelling read for both lovers❤️ & skeptics🤔 of single cell genomics 🧵🧵




Our genome bioinformatics course at EMBL-EBI Training is coming back with a revised programme - this year we go from short- to long-read sequencing! 20-24 Nov 2023, Hinxton, UK Deadline 7th August! Solomon Charles Kayesha Coley Raheleh Rahbari Sean Laidlaw Maxime Tarabichi ebi.ac.uk/training/event…




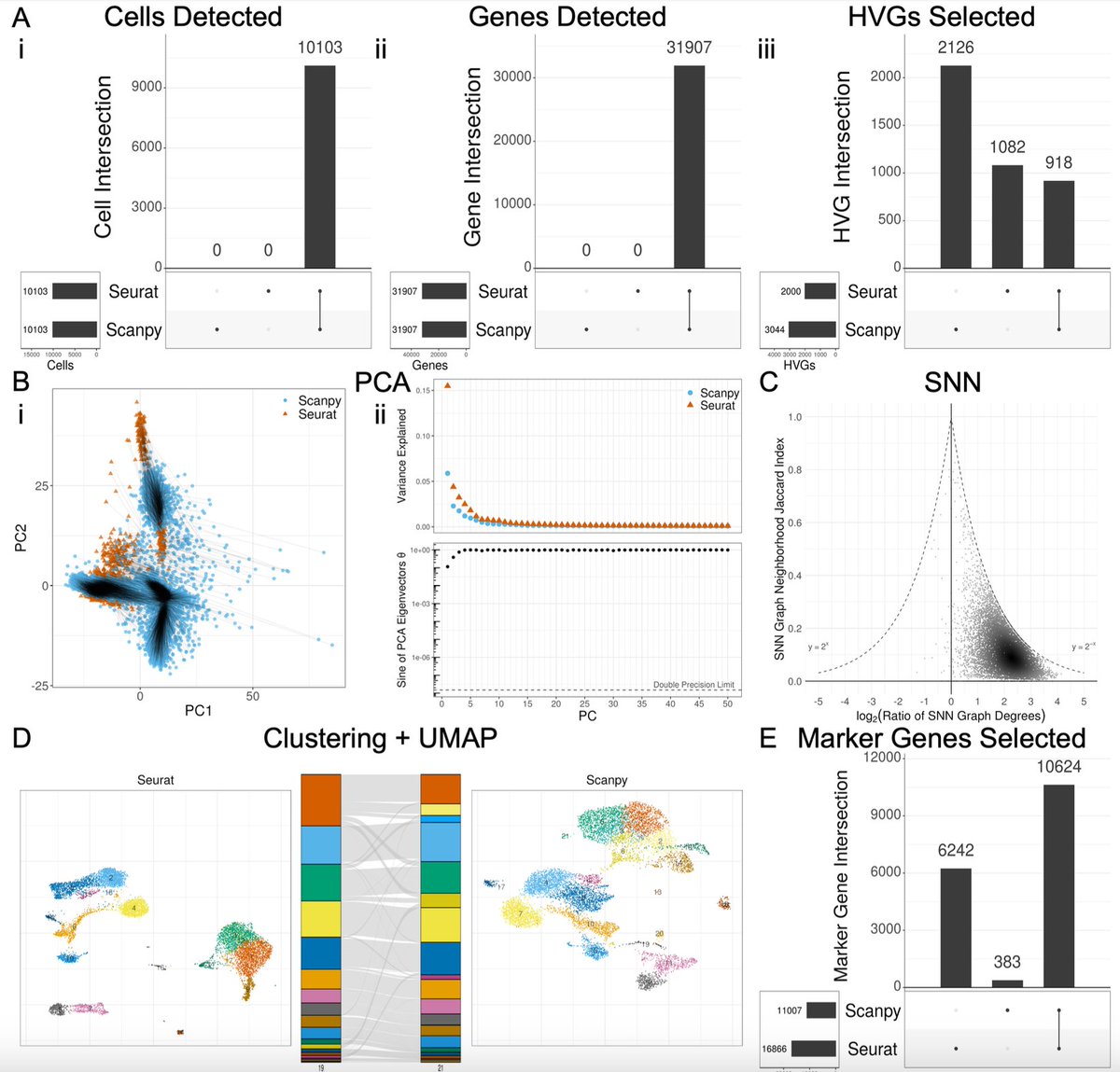
The choice of whether to use Seurat or Scanpy for single-cell RNA-seq analysis typically comes down to a preference of R vs. Python. But do they produce the same results? In biorxiv.org/content/10.110… w/ Joseph Rich et al. we take a close look. The results are 👀 1/🧵





Application deadline for our EMBL-EBI Training sequencing course is 22nd July! It covers all qc and mapping steps, variant calling for short and structural variants, how to analyse both short and long read sequencing data, and the basics of how to build a pipeline. Spread the word!

Stellar team of organisers and trainers including Raheleh Rahbari Pille Hallast Kayesha Coley Sean Laidlaw Maxime Tarabichi Solomon Charles Vic Flores 🦊 and Erik Garrison for our closing keynote lecture!