
Christoph Hahn
@c__hahn
Biology, Bioinformatics | husband & father | curious, fascinated, frustrated, exhausted, excited, hungry, surprised ..
ID: 2843825735
https://github.com/chrishah 25-10-2014 15:40:27
1,1K Tweet
254 Followers
195 Following

How do you stress the importance of replicates to improve the accuracy of #eDNA sampling? Use pizza of course 🍕🍕🍕 A great talk by Nathan Griffiths #fishimp2023



The small variant calling benchmarking initiative of @NGSCN1 is published. Thanks a lot to all the contributors and DFG public | @[email protected] for the funding. (1/x)


Our CARD pilot paper with Kimberley Billingsley Cornelis Blauwendraat @BenedictPaten and many others is online! We establish a wet + dry lab protocol for single nanopore flow cell sequencing, which generates state-of-the-art SNP and SV calls + assemblies. rdcu.be/dl901



Interested in #phylogenetics using whole #genome/#transcriptome/#proteome data? Christoph Hahn and I are very happy to share phylociraptor with you: It facilitates explorative and reproducible phylogenomic analyses of thousands of genes and genomes. Preprint: tinyurl.com/yc4f7v6d
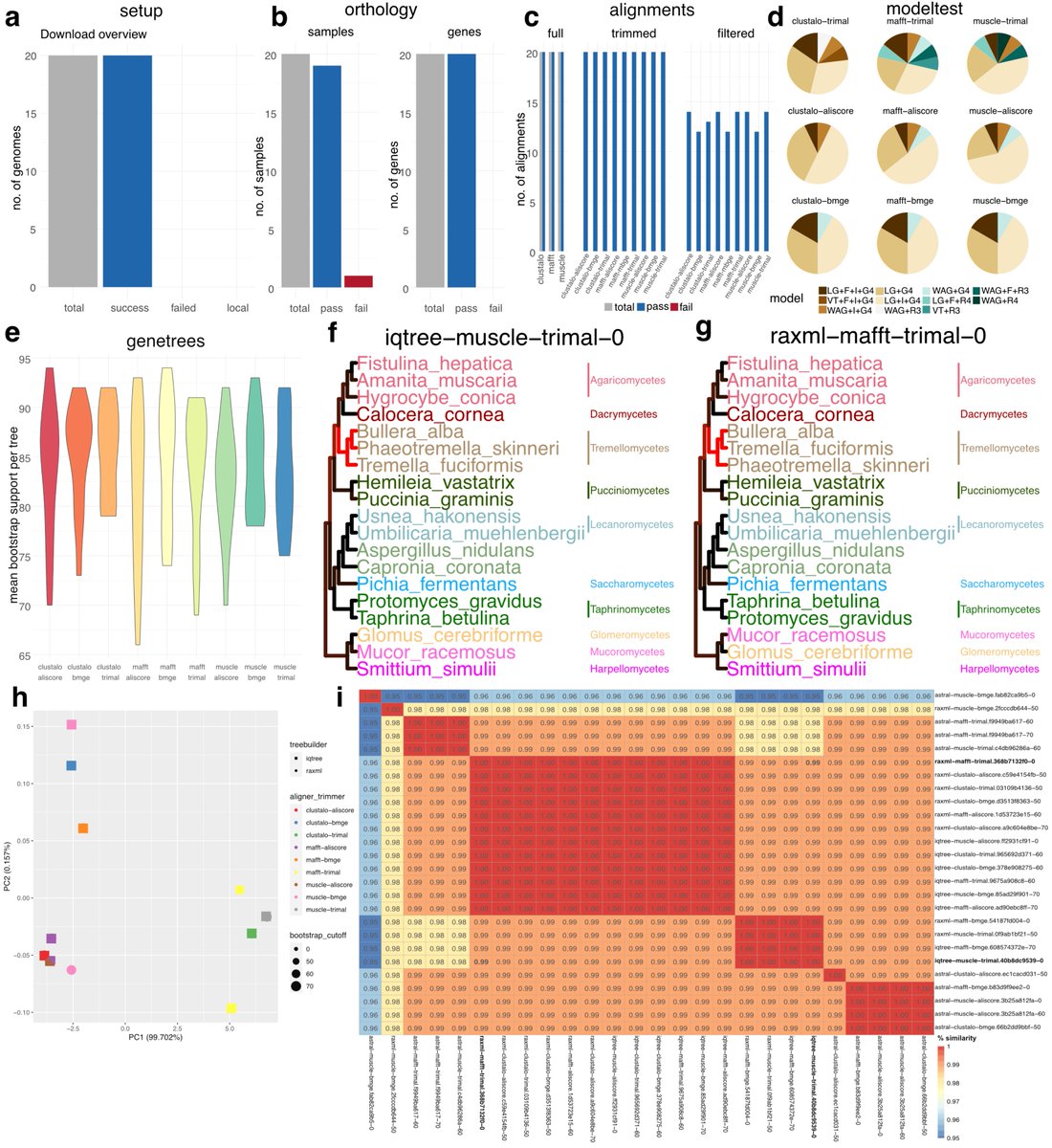

Christoph Hahn Available on GitHub: github.com/reslp/phylocir…


Indeed, thanks Philipp Resl, EcoEvoGraz, Uni Graz/University of Graz. Check it out - even if you're not interested in #phylogenetics. It's about democratizing complex computational analyses and #reproducibiltiy through #scalability and #portability (#containers)..

Structural #genome annotation with BRAKER & TSEBRA - workshop will take place on the 21st Nov. from 9am to 3pm, registration is free but capacity is limited to 35 participants - so hurry! cognitoforms.com/ERGAEuropeanRe… #training #bioinformatics #genomics #biodiversity European Reference Genome Atlas ERGA


Great news from COPO! Reliably brokering sample #metadata is an essential step of the reference #genome production workflow. Congratulations on this impressive milestone 🎉 Biodiversity Genomics Europe (BGE) Earlham Institute



Our preprint is out! We hacked the Oxford Nanopore sequencer to read amino acids and PTMs along protein strands. This opens up the possibility for barcode sequencing at the protein level for highly multiplexed assays, PTM monitoring, and protein identification! biorxiv.org/content/10.110…

Yesterday we released our preprint showing how a Oxford Nanopore array morphs into a protein sequencer... Achieving LONG reads (hundreds of amino acids) and the capability to re-read individual molecules multiple times, all thanks to a protein-processive molecular motor.


