
CambridgeCProteomics
@camcproteomics
ID: 735506809994289152
25-05-2016 16:24:23
36 Tweet
345 Followers
18 Following


Please join us for 12th installment of our EMERGE webinar series featuring Eneko Villanueva Verdejo from CambridgeCProteomics, and hosted by Stoyan Stoychev entitled: RNA-Protein interactions in Space and Time. Registration: us06web.zoom.us/webinar/regist…



Exciting opportunity to join my lab CambridgeCProteomics in a 3 year postdoc position to understand the role of SUMO modification in guiding plant adaptation. jobs.cam.ac.uk/job/38945/ Part of a BBSRC sLoLa project with Ari's lab, Andy Jones, Malcolm Bennett (Nottingham).


A ‘tour-de-force’, as Kathryn would say. A great collaborative project revealing proteomic responses to nearly all genes in S. cerevisiae !! Ralser Lab Georg Kustatscher and many more. Check it out below: cell.com/cell/pdf/S0092…

New E.coli RNA-BP study using OOPS. We show how dynamic these proteins are between different growth phases and find many new RBPs. Great collaboration from Willis Lab MRC Toxicology Unit, Gavin H Thomas labs and CambridgeCProteomics, led by Mie Monti and Eneko Villanueva biorxiv.org/content/10.110…

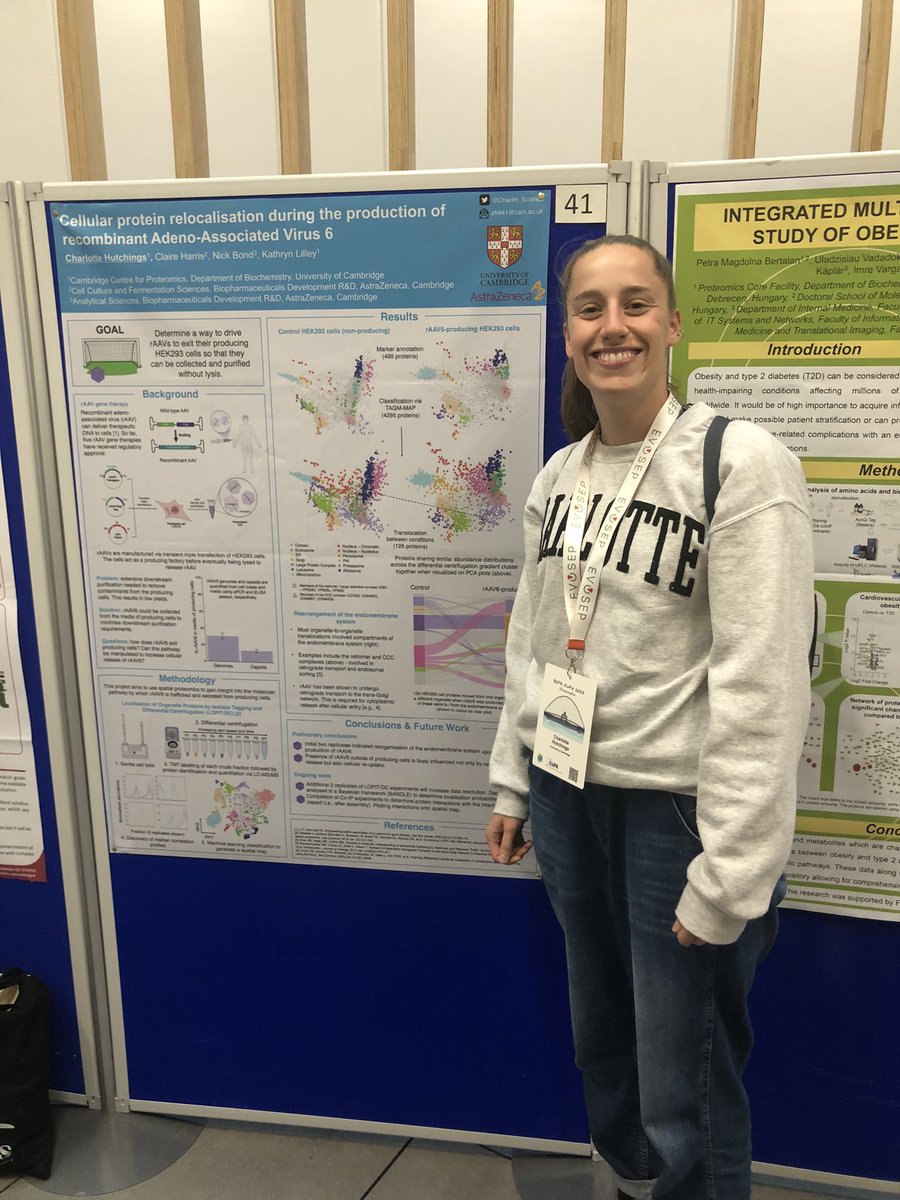
Come visit our PhD students’ posters British Society for Proteome Research #BSPREUPA2023 Kieran is at 135 and Charl at 41 !!


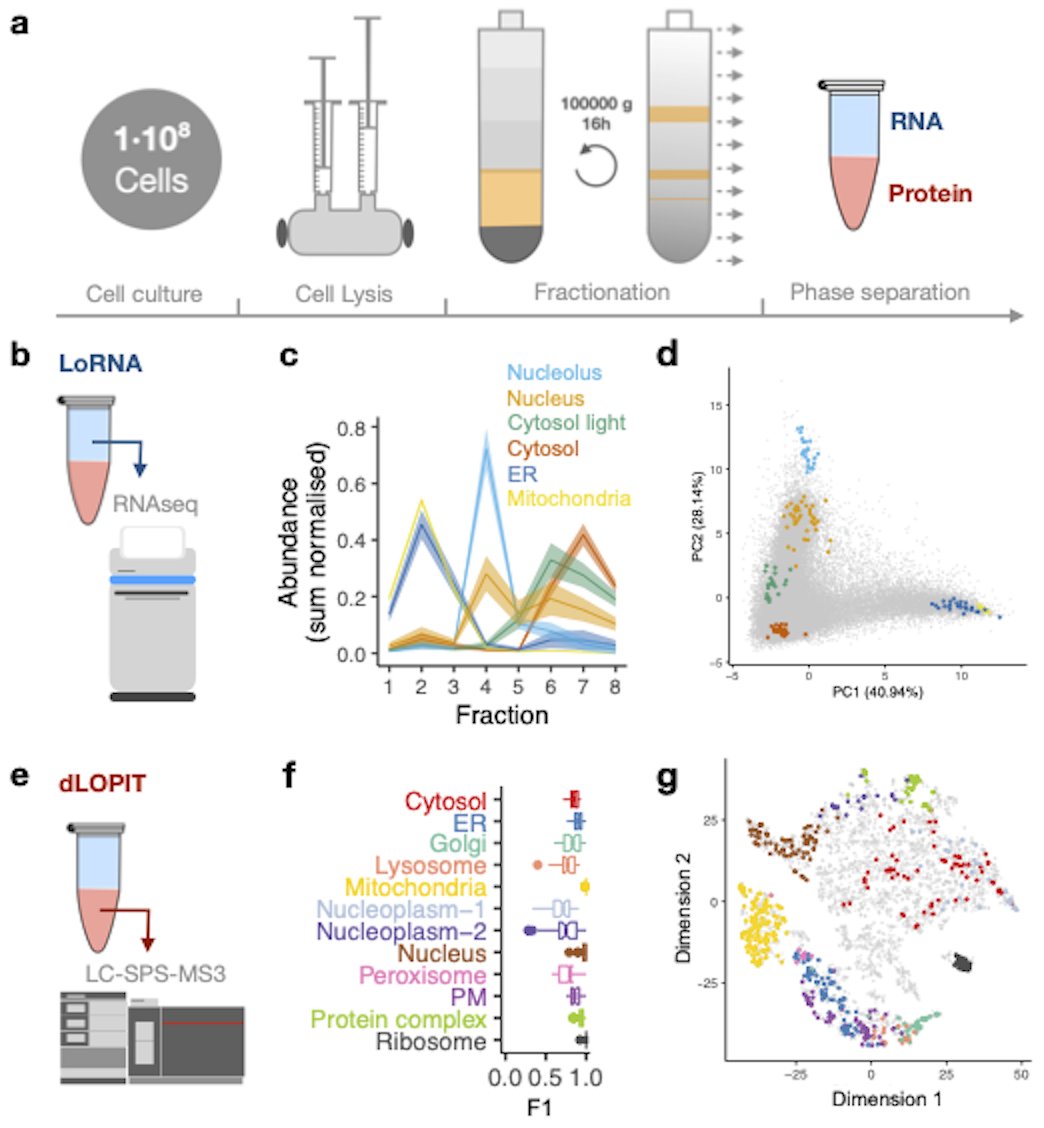
Kathryn Lilley (Kathryn Lilley) and Anne Willis (MRC Toxicology Unit) published a paper in Nature Methods with colleagues Human Technopole and Oxford Statistics that presents the most comprehensive overview to date of subcellular co-localisation dynamics of RNA and protein: bioc.cam.ac.uk/news/new-frame…

Delighted to host Kathryn Lilley CambridgeCProteomics Cambridge Biochemistry as this weeks biomedical seminar speaker Kathryn is presenting her work on the subcellular transcriptome and proteome #transcriptomics #proteomics #omics #biochemistry




#OpenAccess #Review Advances in spatial proteomics: Mapping proteome architecture from protein complexes to subcellular localizations by Kathryn Lilley and colleagues at Cambridge University dlvr.it/TDTn2p



Incredibly honoured to receive the EuPA award. Tremendous thanks to Kathryn Lilley and all amazing colleagues at the CambridgeCProteomics, the birthplace of DIA-NN, as well as my fantastic lab members, our collaborators and the proteomics community for all the support!


Kathryn Lilley (Kathryn Lilley) spoke about her work on cell wide protein localisation to understand therapy resistance mechanisms Cambridge Biochemistry | #BICC2025





