Casper Goverde
@caspergoverde
PhD student | Protein Designer
ID: 1424772011528790019
09-08-2021 16:38:25
43 Tweet
501 Followers
64 Following

We tried to solubilize the claudin, rhomboid, and GPCR membrane folds. Using vanilla ProteinMPNN, we were not able to design soluble versions of these proteins, so Justas Dauparas trained a new version called SolubleMPNN resulted in many soluble designs of these folds.



@ramithuh Casper Goverde Zhidian Zhang Juan José Pierella Turns out Tom Hanks was in town, but, unfortunately, was a bit out of reach.... so Casper Goverde, with the magic of Adobe Photoshop and a little generative ai, made the dream come true 😶🌫️



New amazing research from the Bruno Correia lab! Zander Bühler Harteveld and Alexandra explore protein topologies that have not been found in nature using a novel deep learning pipeline called Genesis




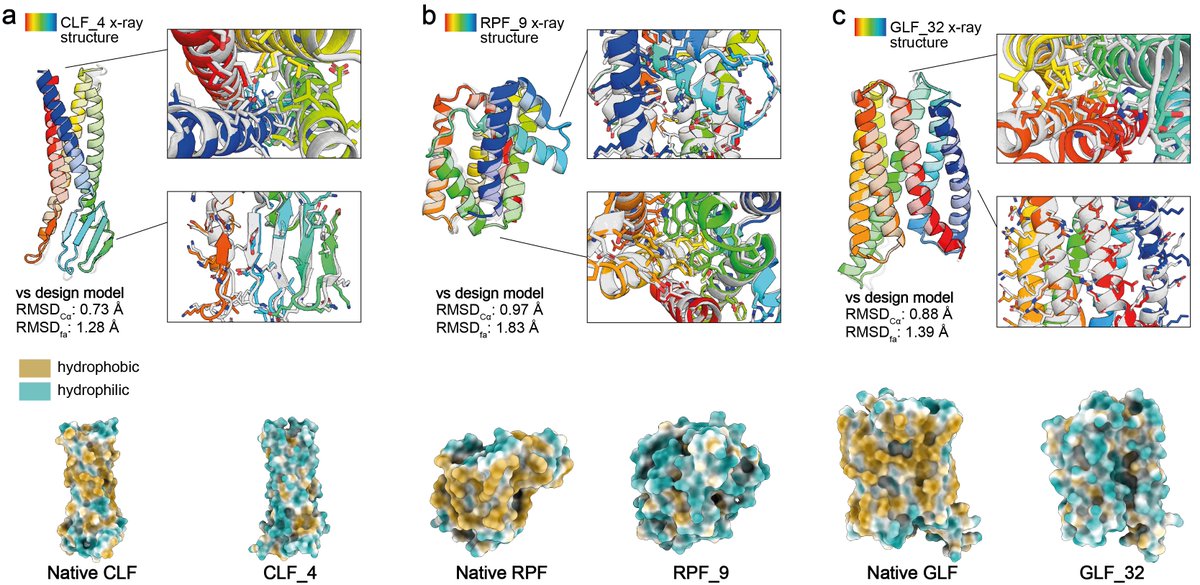
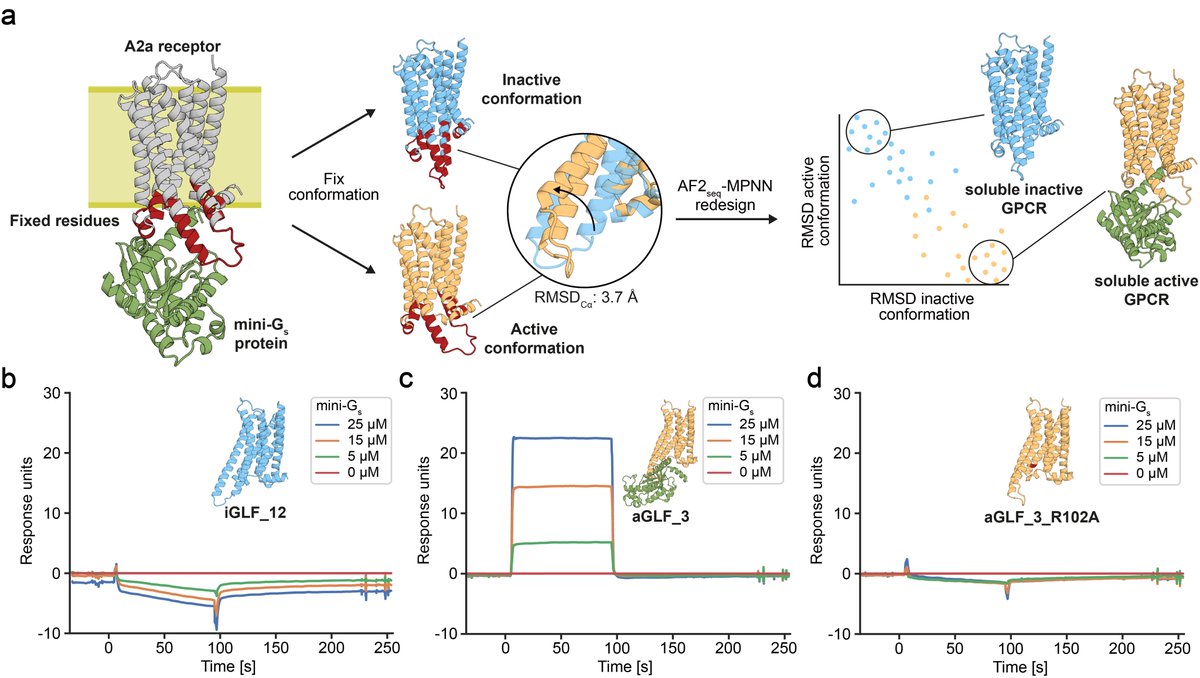
THEY SAID WE COULDN'T DO IT........well, no one actually said that. But in our updated manuscript with Casper Goverde, Nico Goldbach, and Bruno Correia we show you can now design soluble analogues of membrane proteins with preserved functional features! biorxiv.org/content/10.110…

New preprint! In this project spearheaded by Anthony Marchand and Stephen Buckley we designed new chemically-induced protein interactions and experimentally validated three binders targeting Bcl2:Venetoclax, DB3:Progesterone and PDF1:Actinonin complexes. biorxiv.org/content/10.110…

Our method for molecular linker design with diffusion models (DiffLinker) got published in Nature Machine Intelligence 🥳 nature.com/articles/s4225… Thanks a lot to our amazing coauthors Hannes Stärk Clément Vignac Arne Schneuing Víctor Garcia Satorras Pascal Frossard Max Welling Michael Bronstein Bruno Correia

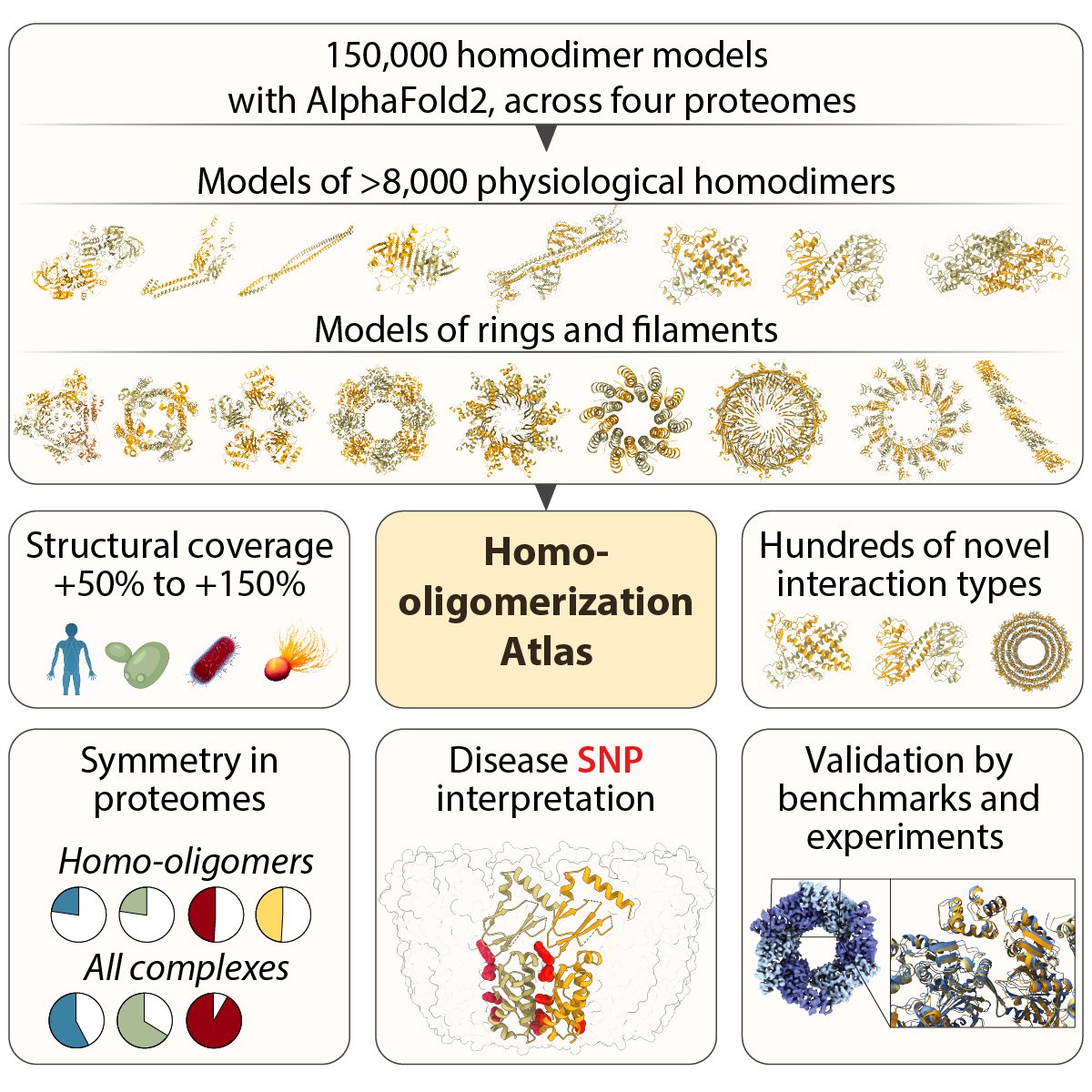
Today in nature we present the results of our exploration of the soluble protein universe and the design of functional soluble membrane proteins with Casper Goverde Nico Goldbach Bruno Correia 1/6 nature.com/articles/s4158…

The last two years of my PhD I've worked with great pleasure with Martin Pacesa Nico Goldbach Bruno Correia and many others. When we started this journey we had no idea where this project would lead us, so I feel deeply humbled that we managed to publish our work in Nature.

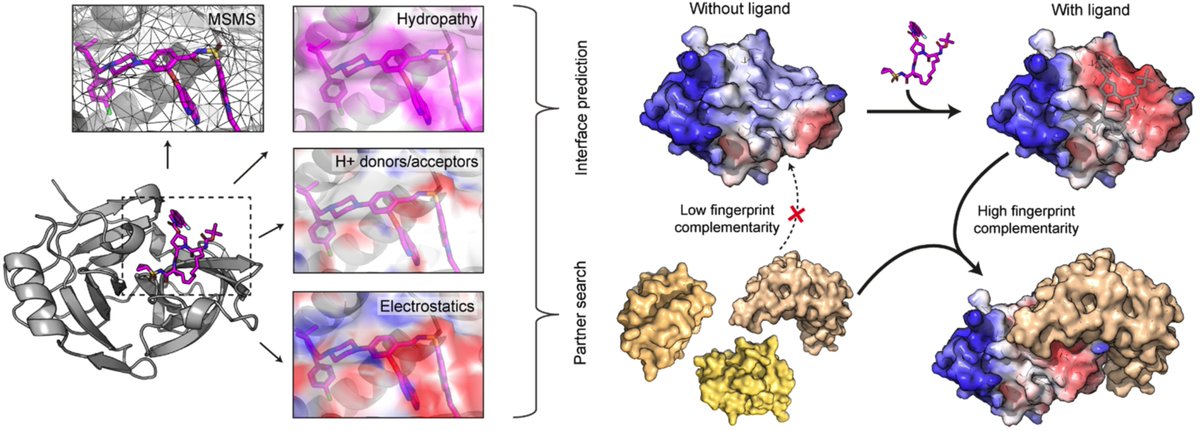
Have you ever wanted to design protein binders with ease? Today we present 𝑩𝒊𝒏𝒅𝑪𝒓𝒂𝒇𝒕, a user-friendly and open-source pipeline that allows to anyone to create protein binders de novo with high experimental success rates. Bruno Correia Sergey Ovchinnikov biorxiv.org/content/10.110…