Charlie Roco
@charlieroco
CTO and cofounder @ParseBio. Co-developer of single cell combinatorial barcoding method SPLiT-seq. Making genomics more scalable, flexible, and accessible.
ID: 791346302386745344
26-10-2016 18:30:35
108 Tweet
1,1K Followers
870 Following

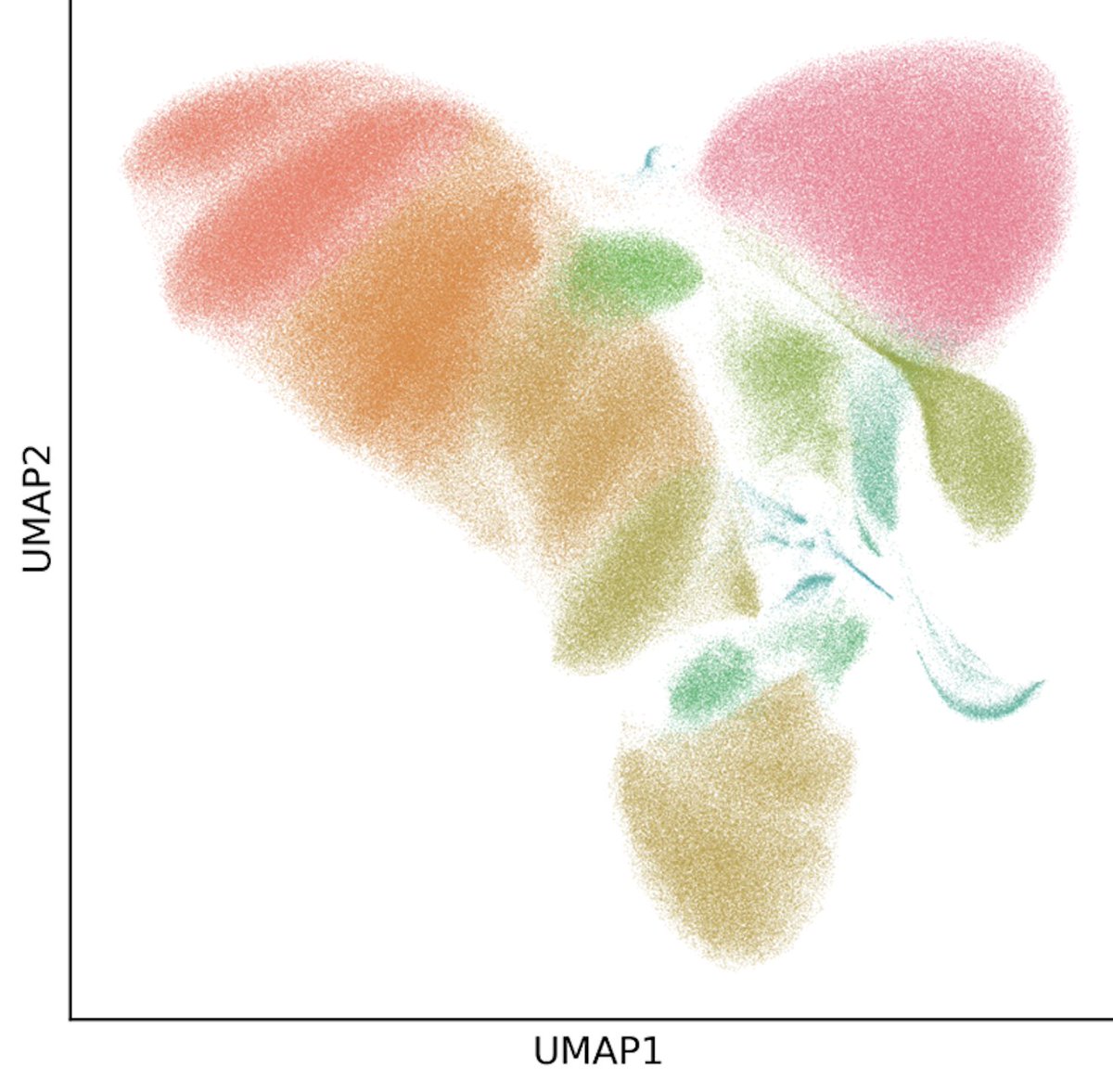
First mega kit through Parse Biosciences and the sub library looks great after sequencing! So convenient to run sub libraries and know what’s going on before the big run. Estimating close to 900k cells for this exp!! Looking forward to the final results but so far so good.🤞🏼

🎙️PeterK & Parse Biosciences will give their talks as part of our #Cajal course on Single #cell profiling and analysis in #neuroscience 📍CGFB 🎟️Free entry Bordeaux Neurocampus Jens Hjerling Leffler ⭐️⭐️🇺🇦⬜️🟥⬜️ The CAJAL Advanced Neuroscience Training Programme PeterK Ana Munoz-Manchado


We had a blast helping to teach a course on #SingleCell profiling and analysis in #neuroscience to the next generation of researchers with #Cajal! The future of single cell is bright! CGFB Bordeaux Neurocampus Jens Hjerling Leffler ⭐️⭐️🇺🇦⬜️🟥⬜️ The CAJAL Advanced Neuroscience Training Programme Ana Munoz-Manchado PeterK






Big thanks to Toon Swings and the entire #RNGS23 organizing committee for inviting me to represent Parse Biosciences in beautiful Ghent! What a great conference - hope to be back again soon!


That was nuclei extraction extraction, using S2 Genomics singulator and fixation w/ Parse Biosciences kits (lucky 13 kits!). Looking forward to the next steps 👩🏻🔬 And how was your week? #postdoclife #biology





Are you attending #ASHG23? Don't miss our featured education session with Rahul Satija (Rahul Satija) from the New York Genome Center. Lunch will be provided for the first 50 attendees.


So excited to work with Parse Biosciences to share my work studying the role of Sox9 in promoting branching morphogenesis in mouse intrahepatic bile ducts (IHBDs). I will highlight how Parse scRNA-seq enabled discovery of a target to rescue IHBD branching in mice lacking Sox9.