
Youngmin Chung
@chungyoungmin
Integrated master & Ph.D. candidate.
Researcher in Next Generation Medicine Lab
Interested in medical deep learning, single cell and spatial transcriptomics.
ID: 1282521869392809985
13-07-2020 03:47:06
337 Tweet
18 Followers
108 Following

Foundation models for single-cell biology should be zero shot, drawing value from emergent organization. We present Universal Cell Embeddings🌌, which can represent cells from any tissue/species in a fixed space Yusuf Roohani @aggwall Jure Leskovec Stephen Quake biorxiv.org/content/10.110…


Excited to share PDGrapher! - a causally-inspired GNN model to predict therapeutically useful perturbagens🧬💊 📜biorxiv.org/content/10.110… 🔬zitniklab.hms.harvard.edu/projects/PDGra… 💫Thank you to amazing collaborators and mentors Marinka Zitnik, Isuru Herath, Kirill Veselkov, Michael Bronstein



Our end-to-end workflow for 3D pathology is now published in Nature Protocols! This includes all the steps to go from archived pathology tissues to 3D H&E-like datasets, with an emphasis on quality control for large studies. Full text at: rdcu.be/dwIYW






New in Nature Methods! We introduce TISSUE, an uncertainty-aware framework for integrating and using #spatial #transcriptomics. Conformal prediction + spatial biology enhances downstream analyses and discovery 💯 Paper: rdcu.be/dyqAj Code: github.com/sunericd/TISSUE 1/3




Is DNA all you need? Introducing Evo, a long context 7B foundation model for biology Evo has SOTA *zero-shot* prediction across DNA, RNA, and protein modalities Evo can generate DNA, RNA+proteins & make CRISPR-Cas systems for first time blog …n-model-tool-arc-institute.vercel.app/news/blog/evo

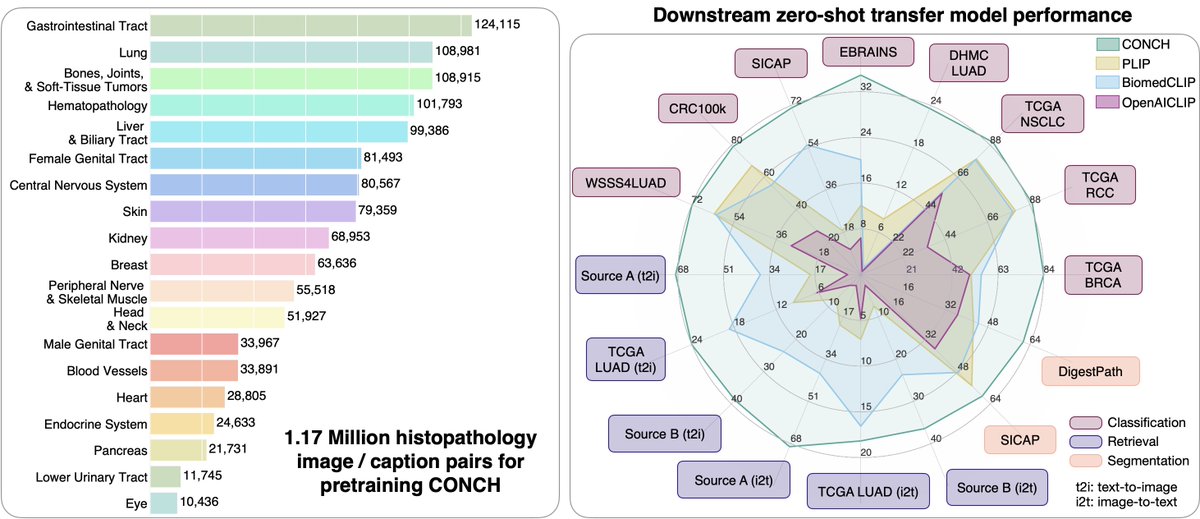
⚡️🔬📣Excited to share our two new Nature Medicine articles, we develop computational pathology foundation models, 1. UNI, a self-supervised computational pathology model trained on 100 million pathology images from 100k+ slides. 2. CONCH, a vision-language model for


⚡️🔬📣Excited to share our new Nature Medicine article, examining disparities in pathology AI models, assessing how modeling choices impact disparities, and evaluating the potential of self-supervised foundation models in mitigating these disparities. nature.com/articles/s4159… See












