Marc Dämgen
@daemgenmarc
Scientist | computational chemistry & AI for drug design | molecular dynamics simulations
ID: 1138456638648700929
11-06-2019 14:43:08
341 Tweet
502 Followers
1,1K Following

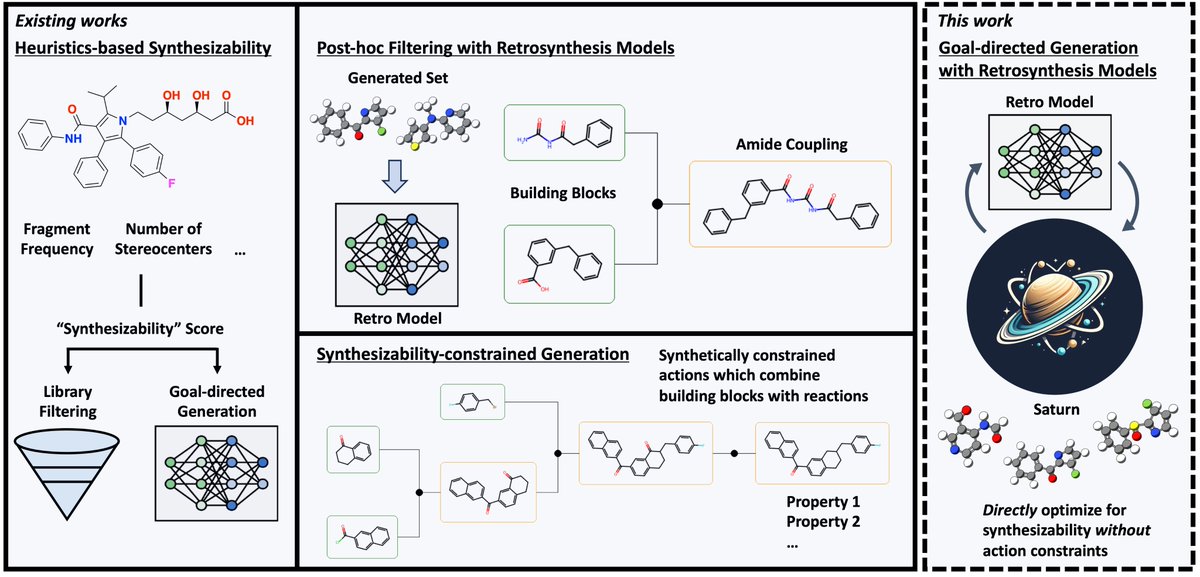
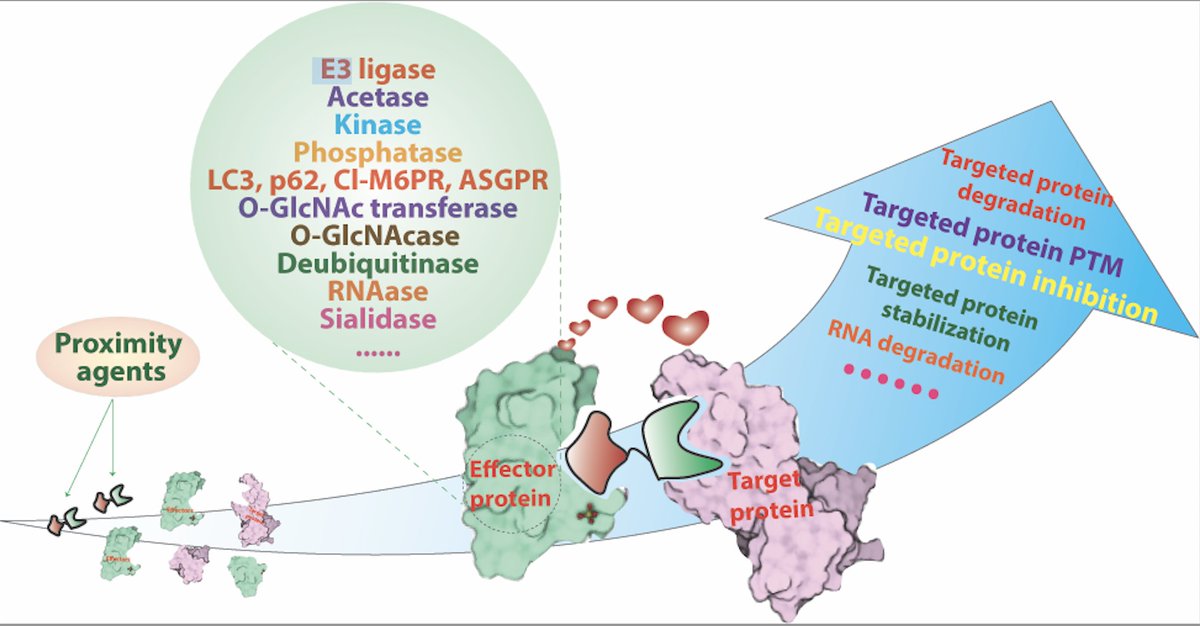
Excited to share our new review on Proximity-Based Modalities for Biology and Medicine | Just out ASAP in ACS Central Science ACS Central Science pubs.acs.org/doi/10.1021/ac…

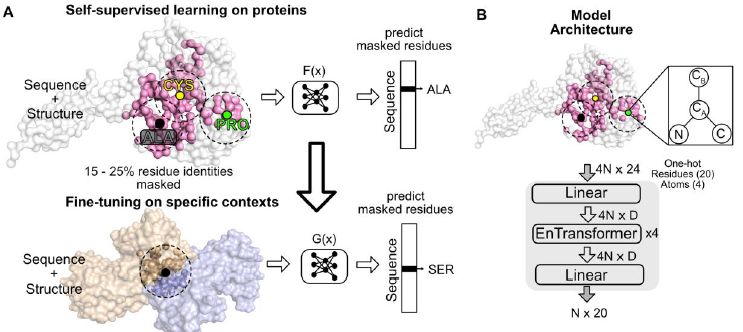
Now out in slightly shorter form :) nature.com/articles/s4158… Kotaro Tsuboyama Justas Dauparas Elodie Laine Sergey Ovchinnikov



Toward docking and interface design, our latest DL tool is a graph neural net for protein interfaces, with the representation learned in different structural contexts. Work by Dr. Sai Pooja Mahajan, also with Jeff Ruffolo. biorxiv.org/content/10.110…

New paper led by Robinson Research Group and first author Haiping Tang. Delighted to have collaborated with the multidisciplinary team via Dheeraj Prakaash and Conrado Pedebos. Fun to work with David B Sauer and Katharina L. Duerr OMass Therapeutics




Thrilled to have our genAI NeuralPLexer research featured on the cover of Nature Machine Intelligence. The pub shows how our model has set the standard for 3D protein-ligand structure prediction for drug discovery. Thanks to our collaborators NVIDIA AI and Caltech nature.com/natmachintell/…




🎉🚀 Excited to share that my internship work, "Benchmarking Active Learning Protocols for Ligand Binding Affinity Prediction," has been published in ACS JCIM & JCTC Journals! 🔗 pubs.acs.org/doi/10.1021/ac… Find 🧵 below for a quick overview. Exscientia






Check out this review article the great Dave Nichols and I wrote on the "Chemistry/structural biology of psychedelic drugs and their receptor(s)". Online today! This is a big milestone as it represents the first independent publication from my lab! bpspubs.onlinelibrary.wiley.com/doi/epdf/10.11…