Dan Polasky
@danpolasky
Research Investigator @ nesvilab - University of Michigan | mass spectrometry, data analysis and software, ion mobility, proteomics
ID: 1145345969501982722
30-06-2019 14:58:53
48 Tweet
235 Followers
102 Following

Love #FragPipe, or only heard about it? If at #ASMS2022, come today (Monday) to our poster 257. Tell us why you like it, what can be improved, or perhaps propose a new collaboration. I will come earlier and will stay as long as people are coming, Fengchao Yu and Dan Polasky also here

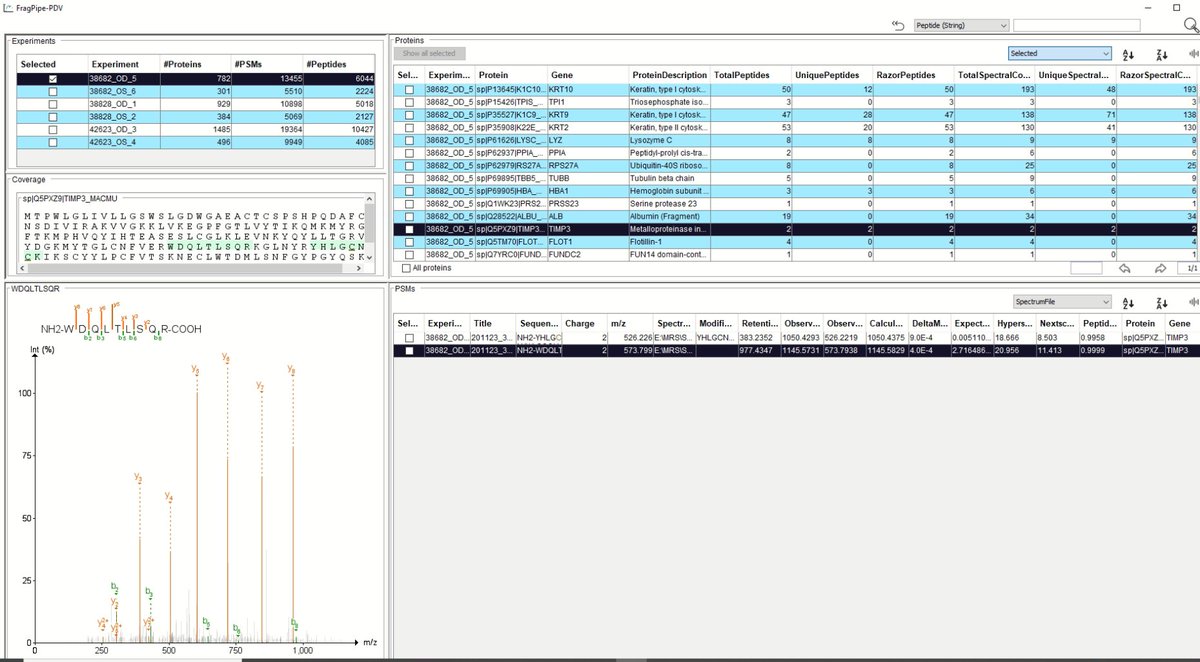
Whoa! I'm really liking Alexey Nesvizhskii Fragpipe 18's new built in spectra viewer! If you have ddaPASEF data it's a great free way to view the PSM's

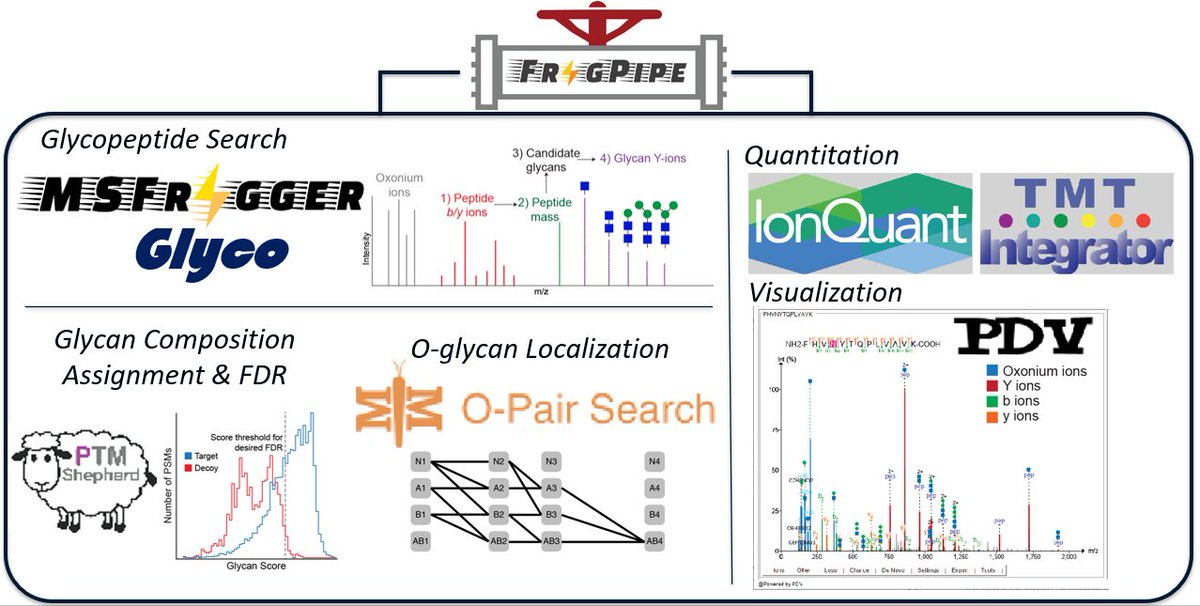
#ASMS2022 come learn about glycoproteomics workflows in #FragPipe and chat with Dan Polasky at his poster ThP187 about MSFragger-Glyco, new glycan assignment with FDR scoring (see recent paper in case you missed mcponline.org/article/S1535-…), quantification, MS/MS spectral viewer, etc

Nice to see our review on glycoproteomics out today in Nature Reviews Methods Primers, it was alot fun putting this together with Stacy Malaker Ieva Bagdonaite Dan Polasky Nick Riley and many more #glycotime

What a great collaboration with the Yifat Merbl lab lab and others, out today in Nature Biotechnology. #MSFragger and #Philosopher are at the core of a computational workflow for PTM mining of immunopeptidome data. We will also be releasing more HLA-PTM workflows in #FragPipe 19 soon.

I am getting an opportunity of a lifetime: I will start as an assistant professor at UWChemistry in the second half of 2023. The Riley Research Group group will explore questions about extracellular biology using mass spec, glycoproteomics, and chemical biology.



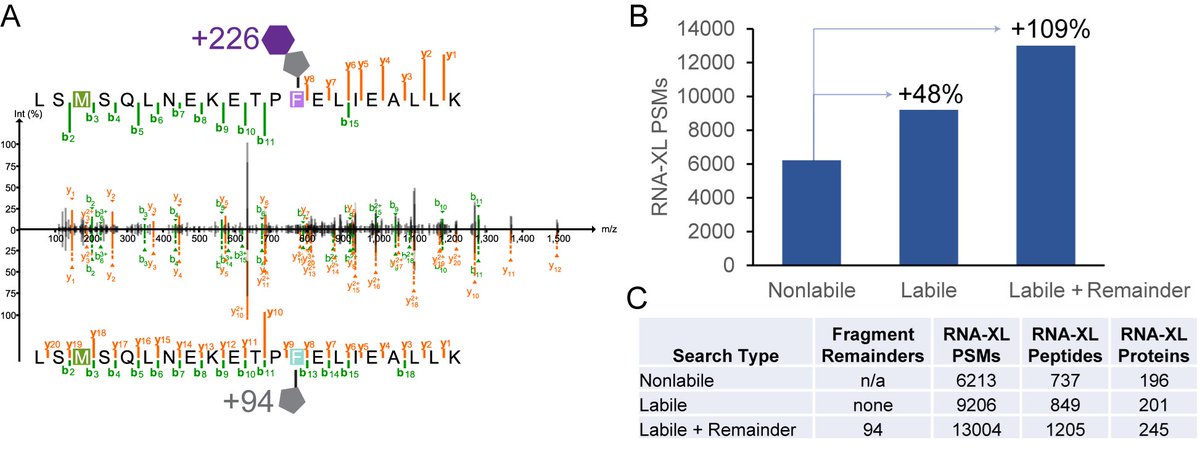
We (led by Dan Polasky) describe a new, flexible Labile Mode of #MSFragger in #FragPipe that can tailor labile PTM searches to their specific fragmentation characteristics. Improved ID rates for phospho, RNA-crosslinked, ADP-ribosylated peptides and more. biorxiv.org/content/10.110…

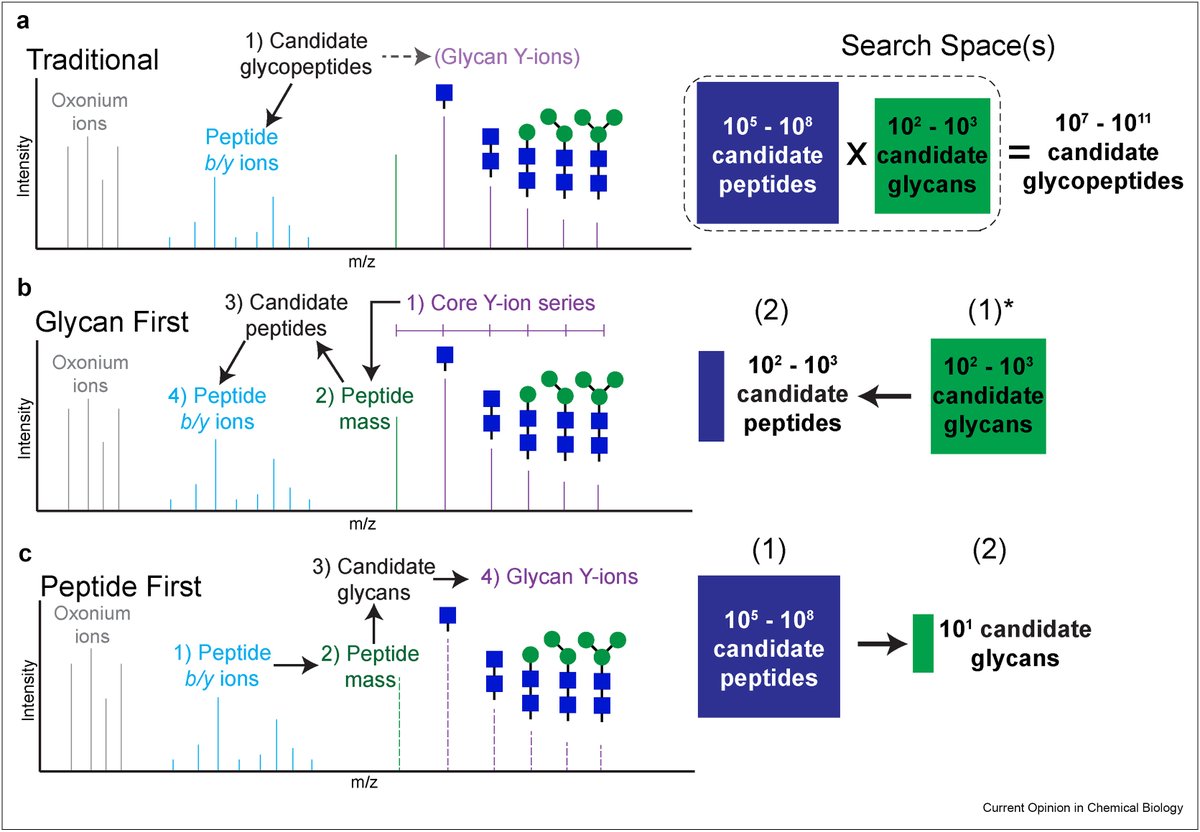
Check out our new review "Recent advances in computational algorithms and software for large-scale glycoproteomics" led by Dan Polasky. We discuss various strategies, new algorithms, and outstanding challenges, including validation of glycan assignments. sciencedirect.com/science/articl…

Dan Polasky is heading to Gordon Research Conference on Glycobiology to present "The MSFragger-Glyco Software Suite in FragPipe: A Growing Toolkit for Glycoproteomics Analysis". Ultrafast search, glyco FDR control, MS/MS visualization, LFQ/TMT quantification, and more! #glycotime

#MSFragger-Labile manuscript led by Dan Polasky is now out in MCP. Many PTMs (phospho, ADP-rybo, etc.) fragment during MS/MS. By incorporating modification fragmentation into search, we can identify many more modified peptides than conventional searches. mcponline.org/article/S1535-…


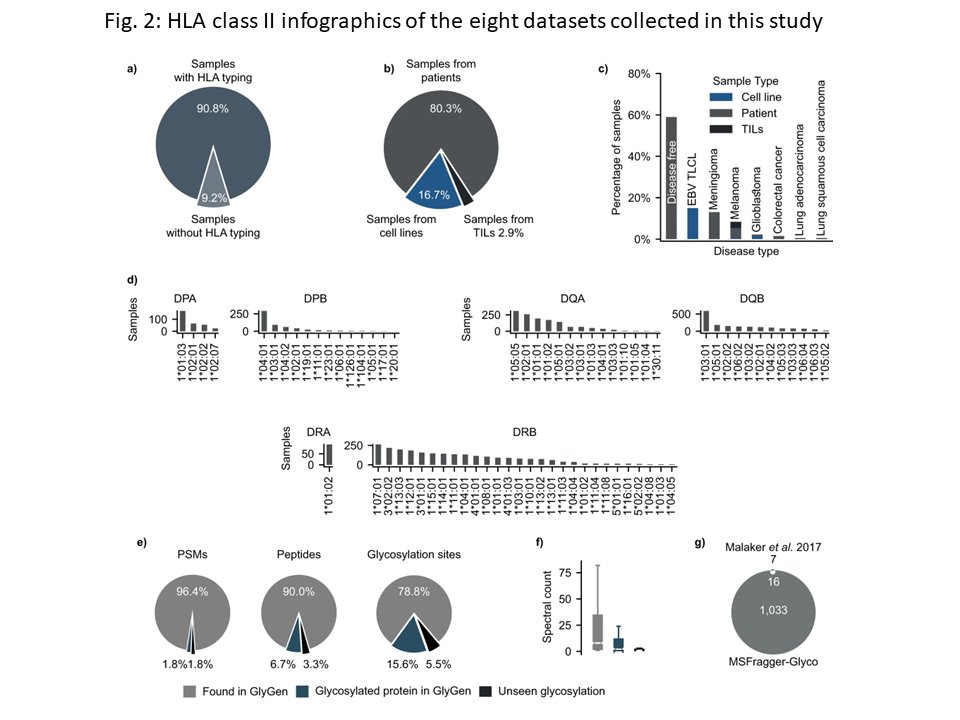
Excited to share our recent work on the glycosylated immunopeptidome: nature.com/articles/s4146… Huge thanks to all the talented team members! Dan Polasky Javier Alfaro Alexey Nesvizhskii #Cancer #Immunotherapy #CancerResearch

Georges BEDRAN, Dan Polasky from Alexey Nesvizhskii and colleagues present a computational workflow for identifying glycosylated peptides from mass spectrometry-based immunopeptidome data and investigate the properties of glycosylated MHC associated peptides nature.com/articles/s4146…


Are you a #FragPipe user attending #HUPO2023 and have a question? Or just want to learn what’s new in MSFragger and other tools (DIA mode, Labile PTMs, MSBooster rescoring, wide window DDA etc). Talk to me, Fengchao Yu or Dan Polasky if you see us and come to our poster presentations




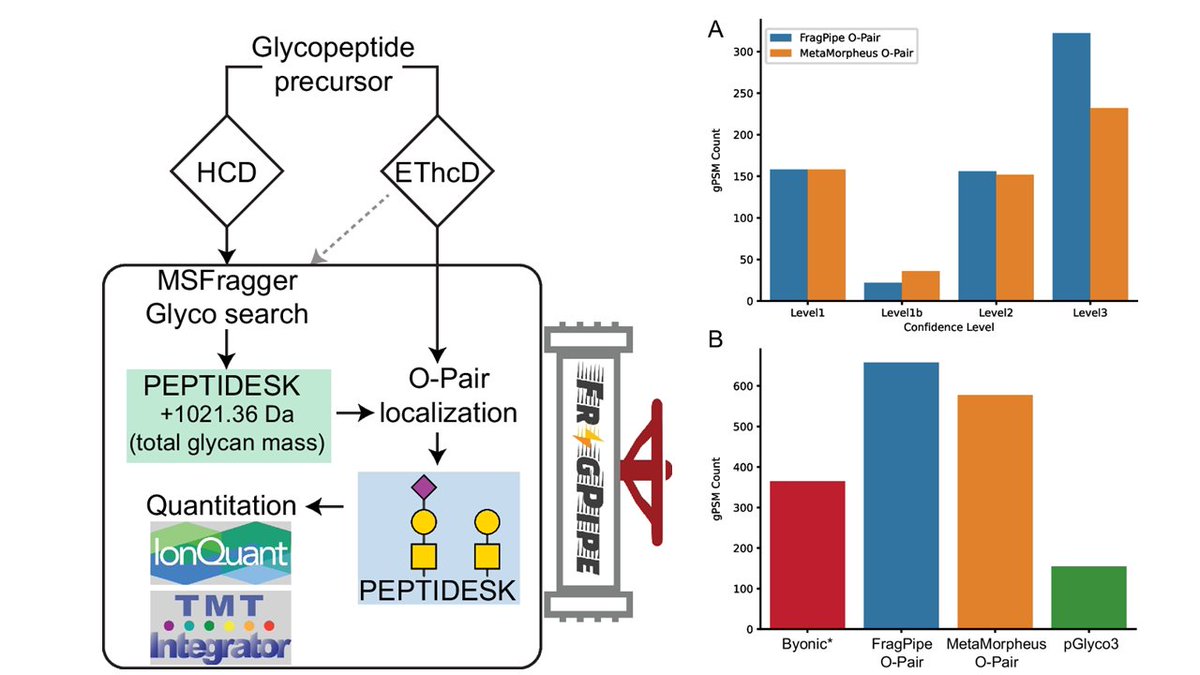
Led by Dan Polasky and in collaboration with MetaMorpheus @metamorpheus.bsky.social lab, we improved #FragPipe for O-glycoproteomics analysis. It provides fast (#MSFragger-Glyco), proteome-wide, quantitative results (LFQ,TMT), and incorporates O-Pair for glycosite localization! link.springer.com/article/10.100…