Daniel Tadros
@danieltadros10
ID: 1242272277980274689
24-03-2020 02:09:35
52 Tweet
46 Followers
68 Following



The devastation in Gaza is unconscionable. We must urge an immediate ceasefire. The killing of Palestinians and Israelis must end. We must also take a hard look at nearly $4 billion a year in military aid to Israel. It is illegal for U.S. aid to support human rights violations.








🎯Check out the paper of Julien Racle & David Gfeller in Immunity. A #machinelearning 🖥️to predict ligands for MHC-II proteins, helping to understand how CD4 #Tcells of the immune system recognize & combat cancer 🦀 cells 🔗bit.ly/42md7Pq Université de Lausanne Ludwig Cancer SIB



Published version of the MixTCRpred paper: rdcu.be/dEKRs. Great work by the fantastic GiancarloCroce . Stay tuned - his next story is coming out shortly 😉.

Predicting MHC-I ligands across alleles and species: How far can we go? biorxiv.org/content/10.110…. Great work by Daniel Tadros Daniel Tadros

Amazing results from huge collaborative efforts with Michal Bassani-Sternberg on #MHC #peptide-binding motifs across a large number of #HLA alleles being presented by David Gfeller at #EFI2024 in Geneva🇨🇭. Data and resources that are fundamental for our applications in #alloreactivity & #HCT!
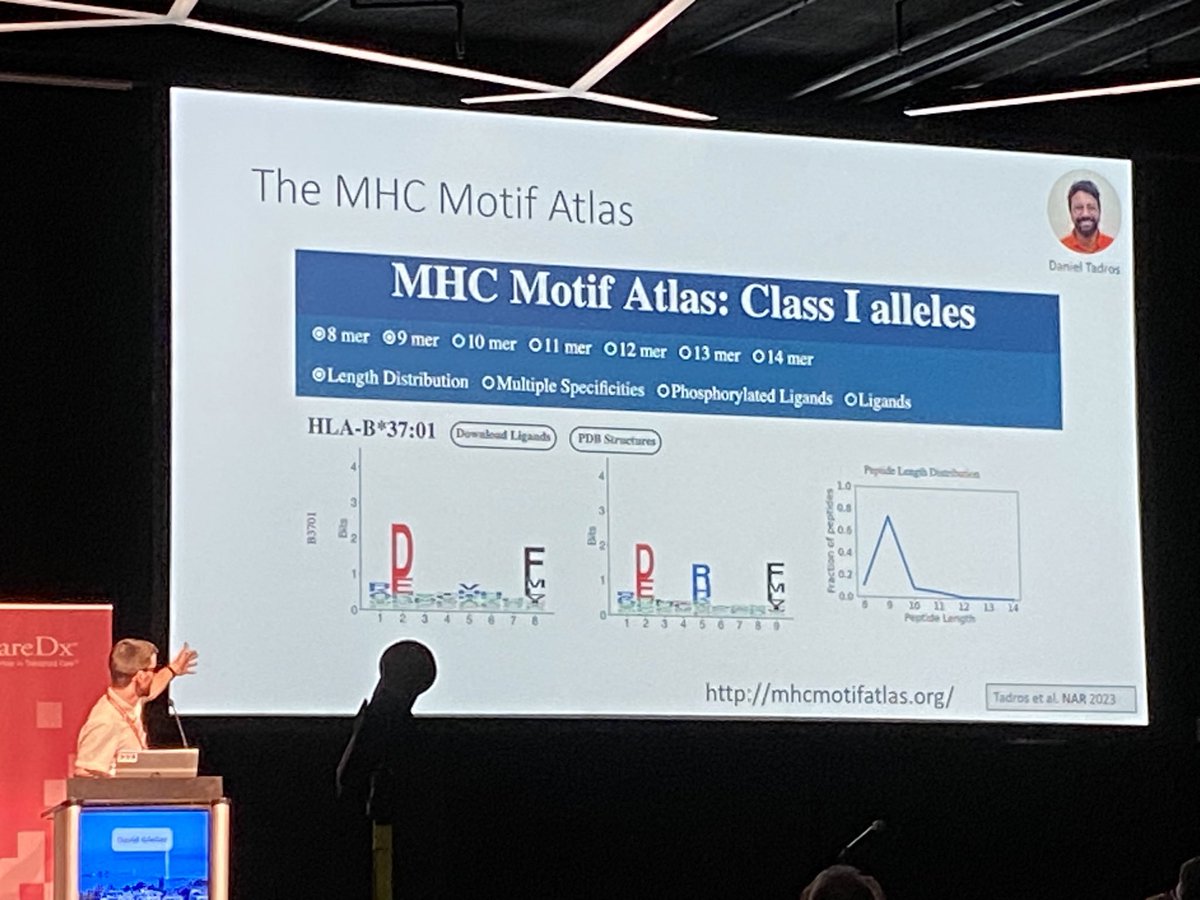

This now complemented beautifully by Julien Racle ‘s data for #HLA class II #immunopeptidomes. Lots of data to feed our immunogenetic questions. Congratulations! EFI_Conference #EFI2025 #EFI2024 David Gfeller Michal Bassani-Sternberg



Our review on the #metacell concept is out in Mol Syst Biol ! Explore our tutorials (github.com/GfellerLab/Met…) and our MetacellAnalysisToolkit (github.com/GfellerLab/Met…) to build and visualize your metacells.

Thrilled to share the beautiful manuscript of GiancarloCroce describing a highly collaborative project with Steven Dunn at CHUV: Phage display profiling of CDR3β loops enables machine learning predictions of NY-ESO-1 specific TCRs biorxiv.org/content/10.110…

Congrats Aurélie Gabriel ✨👏