Fabian
@fabianschuhmann
ID: 1572672903145050112
http://fabianschuhmann.github.io 21-09-2022 19:43:43
42 Tweet
50 Followers
47 Following


Looking forward to 2 days of advanced methods in molecular dynamics simulations Kongelige Danske Videnskabernes Selskab. Talks by Erik Lindahl - eriklindahl.bsky.social, Syma Khalid, Anna Duncan, Max Bonomi, Roberto Covino, Weria Pezeshkian. Organised by @chrisjorg_1, Himanshu Khandelia, Birgit Schiøtt Biomodelling Aarhus Uni & myself mdmeeting2023.wordpress.com


We are happy to introduce our latest program ALISE- the Automated Ligand Searcher, which was made possible because of the great collaborative effort of Fabian Luise Jacobsen Jonathan Hungerland. pubs.acs.org/doi/full/10.10…

Introducing the Automated Ligand Searcher #DrugDiscovery pubs.acs.org/doi/10.1021/ac… Luise Jacobsen Jonathan Hungerland Fabian QuantBio Group #JCIM Vol63 Issue23 #compchem


We used Siewert-Jan Marrink to simulate 700 microseconds of protein dynamics. I'm very glad the study is out. We even won the cover art. Check out the supplementary videos as well :-)

Got back from the DGfB's International Membrane Biophysics Meeting "From Model to Cellular Membranes". A fun meeting, one of the best poster sessions I have attended so far and a pretty location. Three of Weria Pezeshkian's group joined, Neda, Adrià and me (right to left)


Our article was published in #moleculardiversity, available at rdcu.be/dHQOJ! With Felicitas Finke, Jonathan Hungerland, and QuantBio Group, we discuss how receptor structures built with SWISS-MODEL or #AlphaFold behave in #drugscreening for the tick-born #encephalitis virus.

iopscience.iop.org/article/10.108… A very fun project reached its conclusion in this latest published manuscript. With Jonathan and QuantBio Group, we look at and utilize the (surprising) negative eigenvalues in positiv semi-definit matrices :). Check it out!

Yesterday, I presented a talk entitled "Experimental data informs molecular cluster prediction in mitochondria" at ECMTB 2024. Thanks for listening and the picture taken from the audience.




Combining activity assays, molecular dynamics, and networks, we (QuantBio Group,Alice Wong) uncover chemical information pathways across multiple domains in GC-E protein. Important residues match mutation sites that induce various diseases and blindness. Tinyurl.com/pathsgce







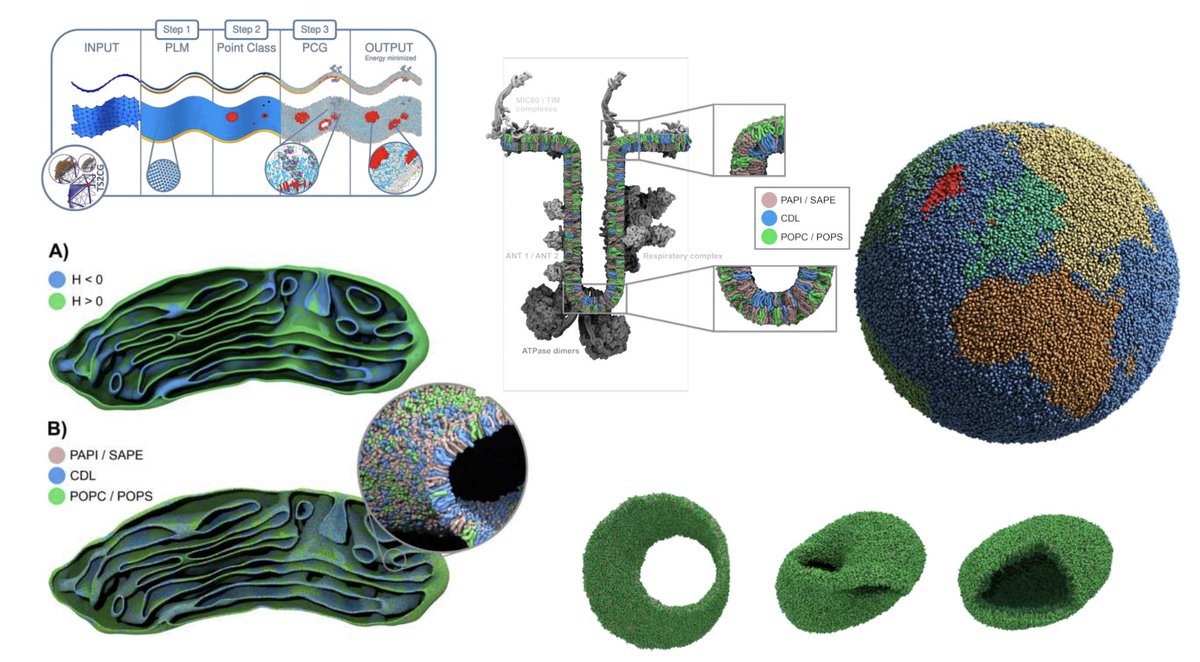
New preprint: TS2CG v2, building and simulating membranes with any shape and lateral organisation. We got Martini Globe, Martini Möbius strip and again mitochondrion. biorxiv.org/content/10.110… Siewert-Jan Marrink Source code: github.com/weria-pezeshki…