FairlieLab
@fairliegroup
The Fairlie group works at the interface of chemistry, biology and disease.
ID: 581493956
http://fairlie.imb.uq.edu.au/index.php 16-05-2012 03:54:25
123 Tweet
272 Followers
21 Following


New research from FairlieLab shows the potential of a new inhibitor combination in the fight against #cancer. #IMB #2020IMB Discover more: bit.ly/2ZKZMDl

New research from FairlieLab shows the potential of a new inhibitor combination in the fight against #cancer. #IMB #2020IMB Discover more: bit.ly/2ZKZMDl

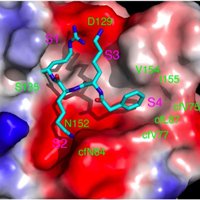
Our new #MR1 study with H McWilliam & J Villadangos, via new tools and techniques incl. a stable fluorescent ligand, conformation specific Ab, and genome-wide CRISPR screen #MAIT #ozchem FairlieLab Institute for Molecular Bioscience @ImagingCoE Doherty Institute Bio21 Institute pnas.org/content/early/…


Pleased to share our new paper - a review on our synthesis and development of ligands that modulate #MR1 and #MAIT cells in Accounts of Chemical Research ACS Publications FairlieLab Institute for Molecular Bioscience @CellsMait #ozchem pubs.acs.org/doi/10.1021/ac…



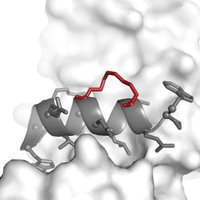
Great to see this paper on stapled peptides out in RSC Chemical Biology. Glad to have played a small part in this work from my time FairlieLab. We investigated how crosslinks affect helicity, protein affinity, proteolytic stability and cell uptake. pubs.rsc.org/en/content/art…

Very happy to share that I have joined Chemistry - University of Strathclyde as a Chancellor's Fellow at University of Strathclyde! Immensely thankful for the support from my mentors: FairlieLab, Odell Group Uppsala universitet, Adam Nelson, BOOMchemistry.com and @patton_lab!




Now we have FairlieLab speaking about strategies for programming peptides for cell permeability and bioavailability If we could reliably get peptides into cells and send them to specific targets, the application possibilities would be endless! #CIPPS2022










