
Genome in a Bottle
@genomeinabottle
The Genome in a Bottle Consortium develops reference materials, reference data, and reference methods needed to benchmark human genome sequencing
ID: 778427430
http://www.genomeinabottle.org 24-08-2012 15:39:49
647 Tweet
3,3K Followers
282 Following

Has anyone made a Genome in a Bottle joke yet? nature.com/articles/s4200… Genome in a Bottle #GIAB

We are proud to announce a new hg38 track group consisting of Human Pangenome Reference Consortium data. In this first release, there are tracks displaying features and variations of HPRC results compared to hg38, as well as alignment tracks. See the full announcement: bit.ly/HPRCdataInUCSC…




Preprint on Exploring gene content with pangenome gene graphs: arxiv.org/abs/2402.16185. It describes pangene for building gene graphs and for calling gene-level variations which can be found at pangene.bioinweb.org. Pleasant collaboration with Maximillian Marin and Maha Farhat.


Our 10th LR seminar BCM HGSC at 22nd March 10am CT! Come join us and listen to new advancements on LR from Miten Jain Tychele Turner Genome in a Bottle Vanessa Porter, PhD Register: hgsc.bcm.edu/LongReadSeminar BCMHouston BCM Department of Molecular and Human Genetics PacBio Oxford Nanopore Rice Computer Science Texas Medical Center



Want to learn more about the tricks and tips for generating ONT Ultra-long DNA? The T2T Consortium and UCSC SeqTech Center are hosting a Technology-focused Webinar next Thursday with a group of experts. Register today: bit.ly/3PFTa2f Adam Phillippy


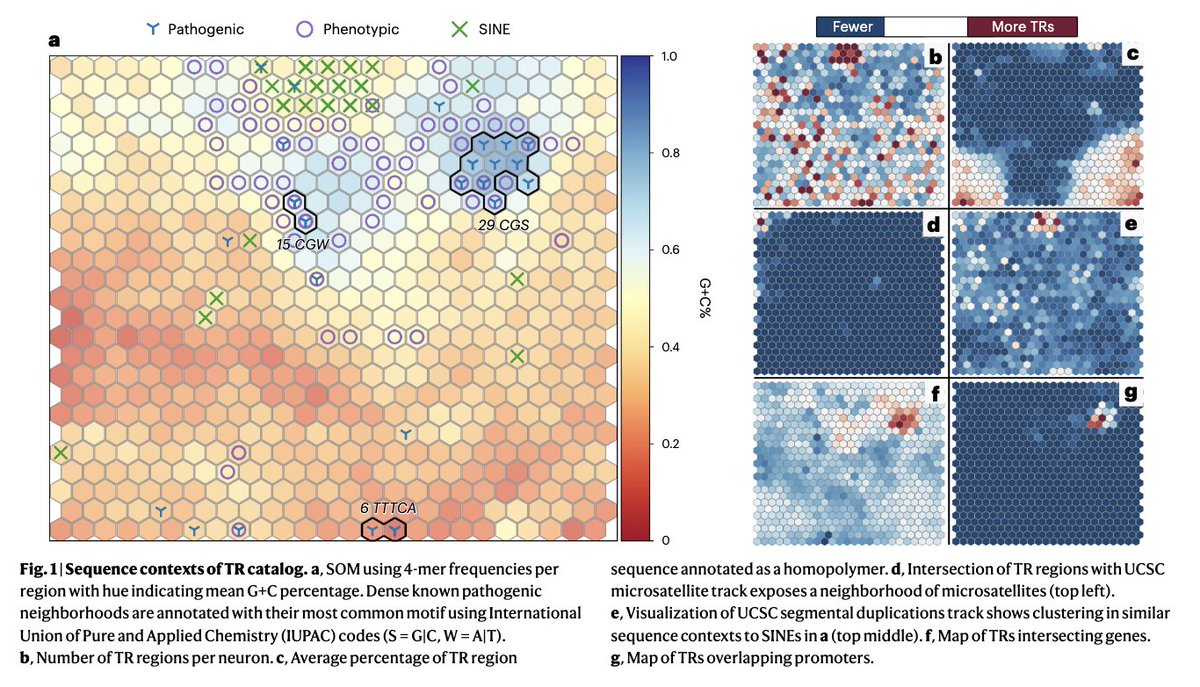
Tandem repeat Genome in a Bottle benchmark Nature Biotechnology out today: rdcu.be/dFQNN . Characterization of 1.7 million TR + benchmark variants + new method to overcome var. representation issues! Great work lead by Adam E. BCM HGSC & great collab. from so many! (1/4)


Extensive WGS for 1st National Institute of Standards and Technology GIAB tumor cell line, consented for public genomic data doi.org/10.1101/2024.0…. PDAC tumor/normal pair with data from Oxford Nanopore PacBio's HiFI&Onso Illumina Element Biosciences Ultima Genomics BioSkryb Genomics, Inc. Arima Genomics Phase Genomics Bionano Kromatid Inc.



We realized they could be mosaic variant positions. Fortunately, NIST Genome in a Bottle released a set of annotated mosaics in the cell lines we were looking at. Plotting those examples revealed they overlap the cluster we inspected.


The Genome in a Bottle genomic stratifications resource is published in Nature Communications: nature.com/articles/s4146… Stratifications reveal key insights into precision and recall of variant calling across different genomic contexts! Great team work by Nate Dwarshuis, Justin Zook & others


don't use hg38.fa as-is. checkout the references 😜 here: ftp-trace.ncbi.nlm.nih.gov/ReferenceSampl… rendered the ipynb (not mine) here: gist.github.com/brentp/1935e9b… in short, use: GRCh38_GIABv3_no_alt_analysis_set_maskedGRC_decoys_MAP2K3_KMT2C_KCNJ18.fasta.gz other updates on the best hg38 reference?

We are pleased to announce the release of the Genome in a Bottle (Genome in a Bottle) Problematic Regions tracks for the hg38 and hs1 human assemblies. Learn more about the release from the following news post: genome.ucsc.edu/goldenPath/new…

Our first curated draft somatic structural variant benchmark for the new GIAB PDAC tumor cell line HG008-T is at ftp-trace.ncbi.nlm.nih.gov/ReferenceSampl…, based on extensive short+long read sequencing data described in doi.org/10.1101/2024.0…. Feedback to improve future versions is very welcome!

Nice to see the impact of this important work. This benchmark set (v4.2.1) drove a huge amount of improvement sequencing. All seq instruments use it to quantify performance. Best credit is to Genome in a Bottle, who drove the field forward both in this work and over a decade.

