
Georges BEDRAN
@georgesbedran3
ImmunoOncology
ID: 1143772101372981248
26-06-2019 06:44:54
53 Tweet
61 Followers
165 Following

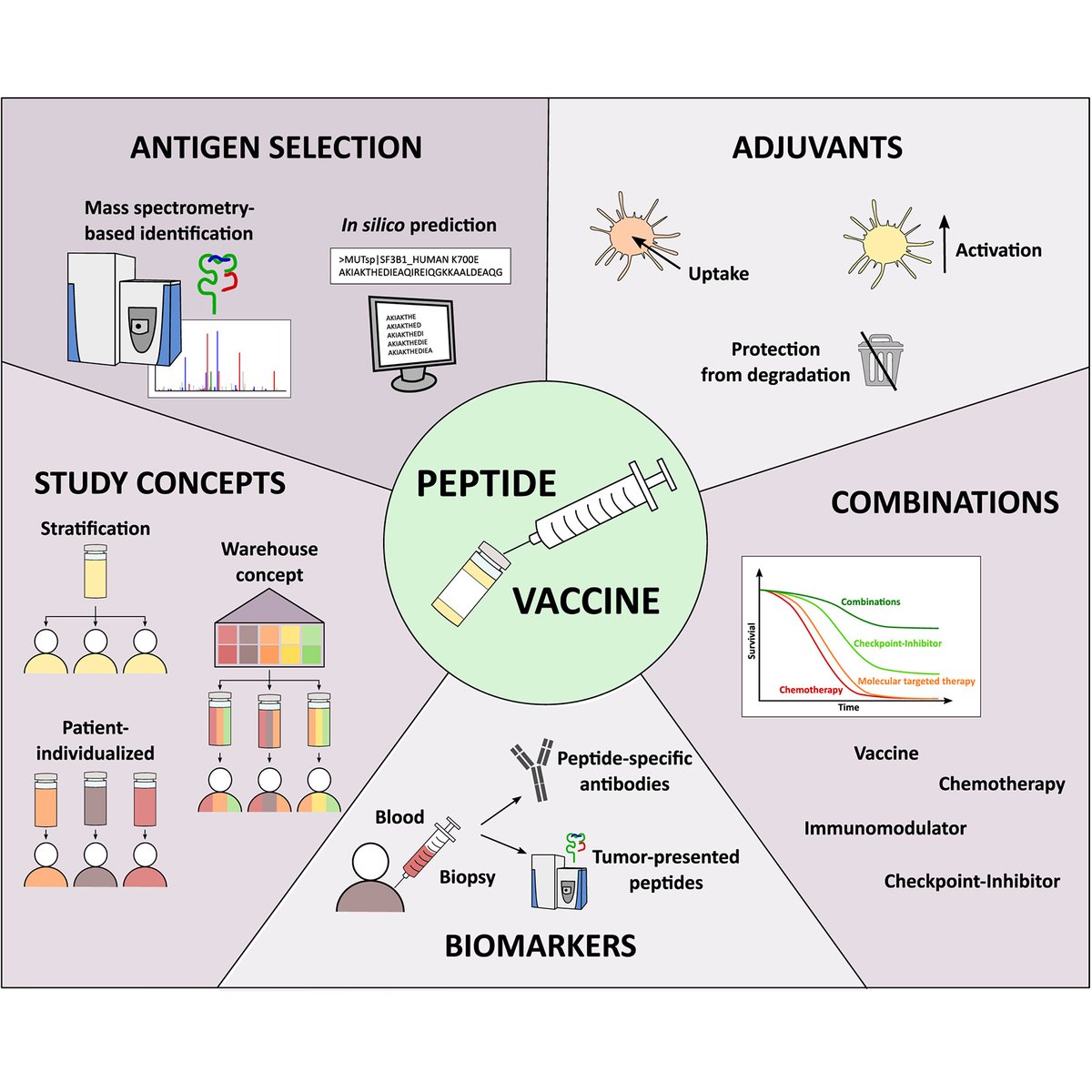
"The Peptide Vaccine of the Future" Nelde et al. of Universität Tübingen discuss current approaches in the development of peptide-based vaccines and critical implications for optimal #vaccine design ― Annika Nel Peptide-based Immunotherapy #Immunopeptidomics #Immunology #TeamMassSpec mcponline.org/article/S1535-…


A small fraction of researchers in KATY #WorldCancerDay Alexander Laird RVCrypto Yamir Moreno Christophe Battail Catia Pesquita Francisco Lupiáñez-Villanueva Laurent Guyon Javier Alfaro @kbarud Georges BEDRAN Aria Nourbakhsh Michele Mastromattei


Thrilled to share our Pan-Cancer Proteomic Map of 949 Human Cancer Cell Lines, now on bioRxiv. A huge collaboration between CMRI #ProCan and Garnett Lab Wellcome Sanger Institute Cancer Dependency Map Sanger. Thanks to @emanuelvgo, Simon Cai and the whole team. biorxiv.org/content/10.110…


I’m delighted to share our Nature Cancer paper by Anne Kraemer et al. nature.com/articles/s4301… Our objective was to investigate whether there are substantial differences in the antigenic landscape presented in tumors that are T cell inflamed versus those that are non-inflamed.




Georges BEDRAN, Dan Polasky from Alexey Nesvizhskii and colleagues present a computational workflow for identifying glycosylated peptides from mass spectrometry-based immunopeptidome data and investigate the properties of glycosylated MHC associated peptides nature.com/articles/s4146…


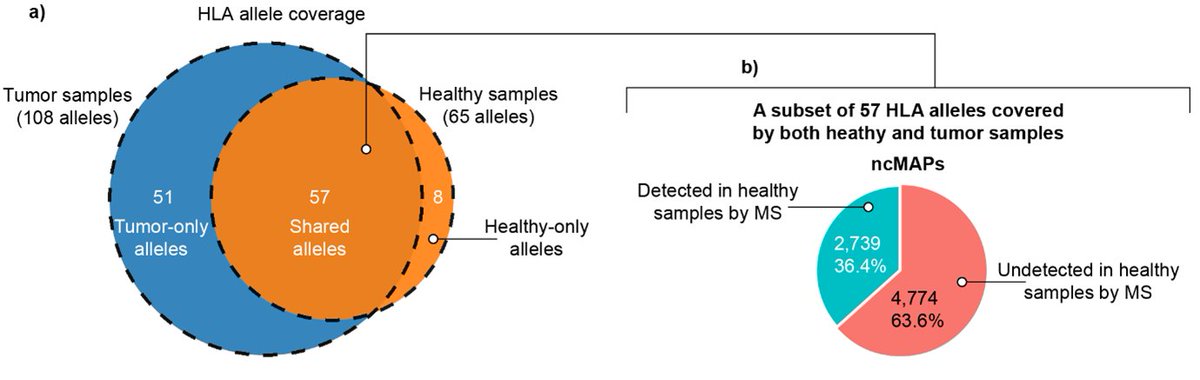
An atlas of non-canonical MHC class I–associated peptides is created and provides potential targets for #Tcell therapies or vaccines— In this study by Georges BEDRAN, Sachin Kote, Javier Alfaro et al. bit.ly/3PiCEWG Dominika Bedran Neuroscientist Fabio Massimo Zanzotto Aleksander Pałkowski goodlettlab


Led by Fengchao Yu we present MSFragger-DIA for direct peptide identification from DIA data. It leverages the speed of fragment ion indexing and searches DIA MS/MS spectra prior to feature detection and peak tracing. Complete DIA analysis workflow in #FragPipe nature.com/articles/s4146…

MSBooster manuscript led by Kevin Yang is out! In tandem with Percolator, it improves identification rates in #FragPipe using deep learning-based predictions (from DIA-NN). We illustrate it using HLA peptidomics, direct DIA, single-cell, and timsTOF data. nature.com/articles/s4146…


