Haotian Cui
@haotiancui1
Building virtual cell. Large models for drug discovery. CS Ph.D. @UofT.
ID: 1169329330369060872
https://subercui.github.io/ 04-09-2019 19:20:21
57 Tweet
206 Followers
503 Following





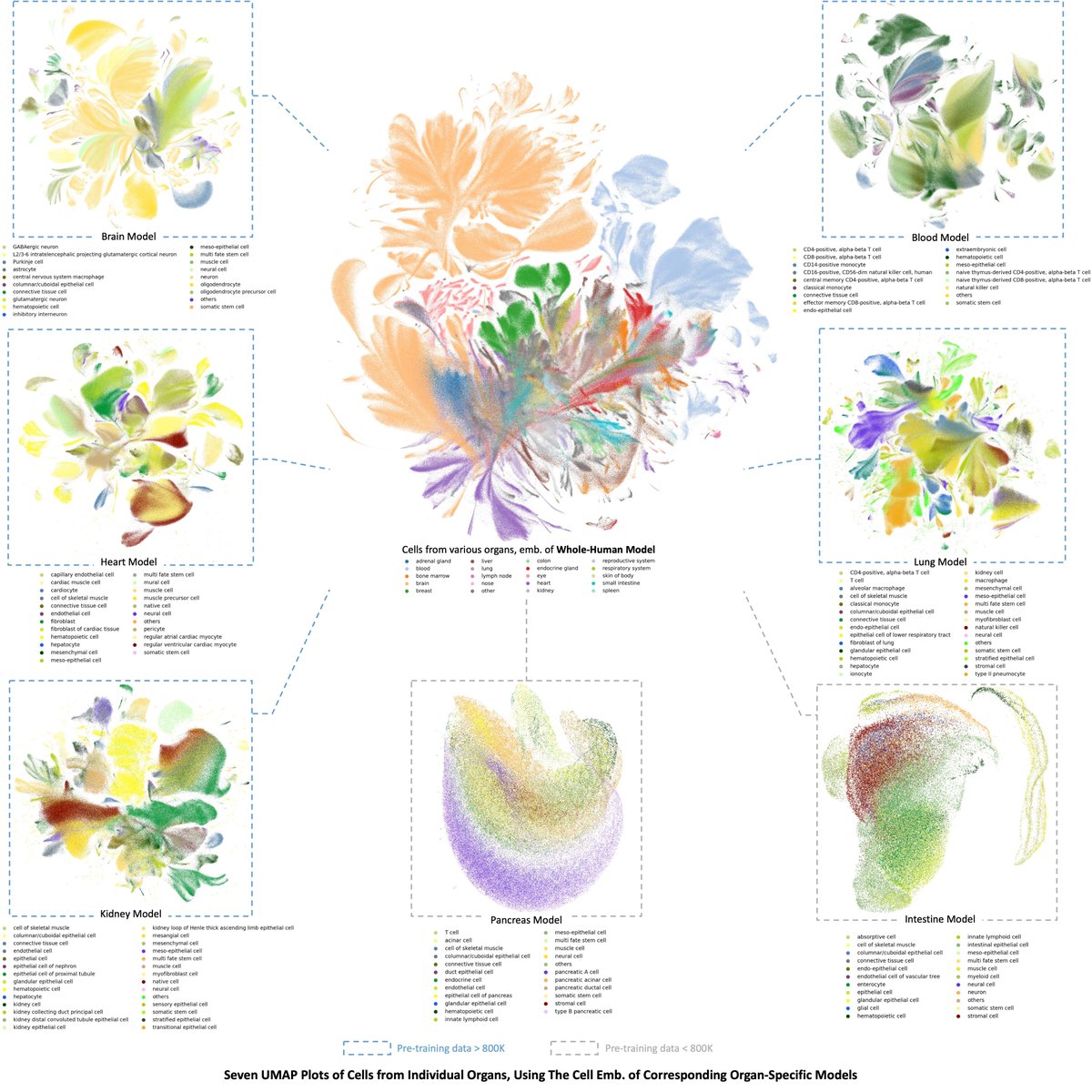
Glad and super excited about this collaboration with #CELLxGENE. Now you can access scGPT embeddings directly from the census API online, and enjoy zero-shot reference mapping and other tasks seamlessly @CZIScience Human Cell Atlas Jonah Cool





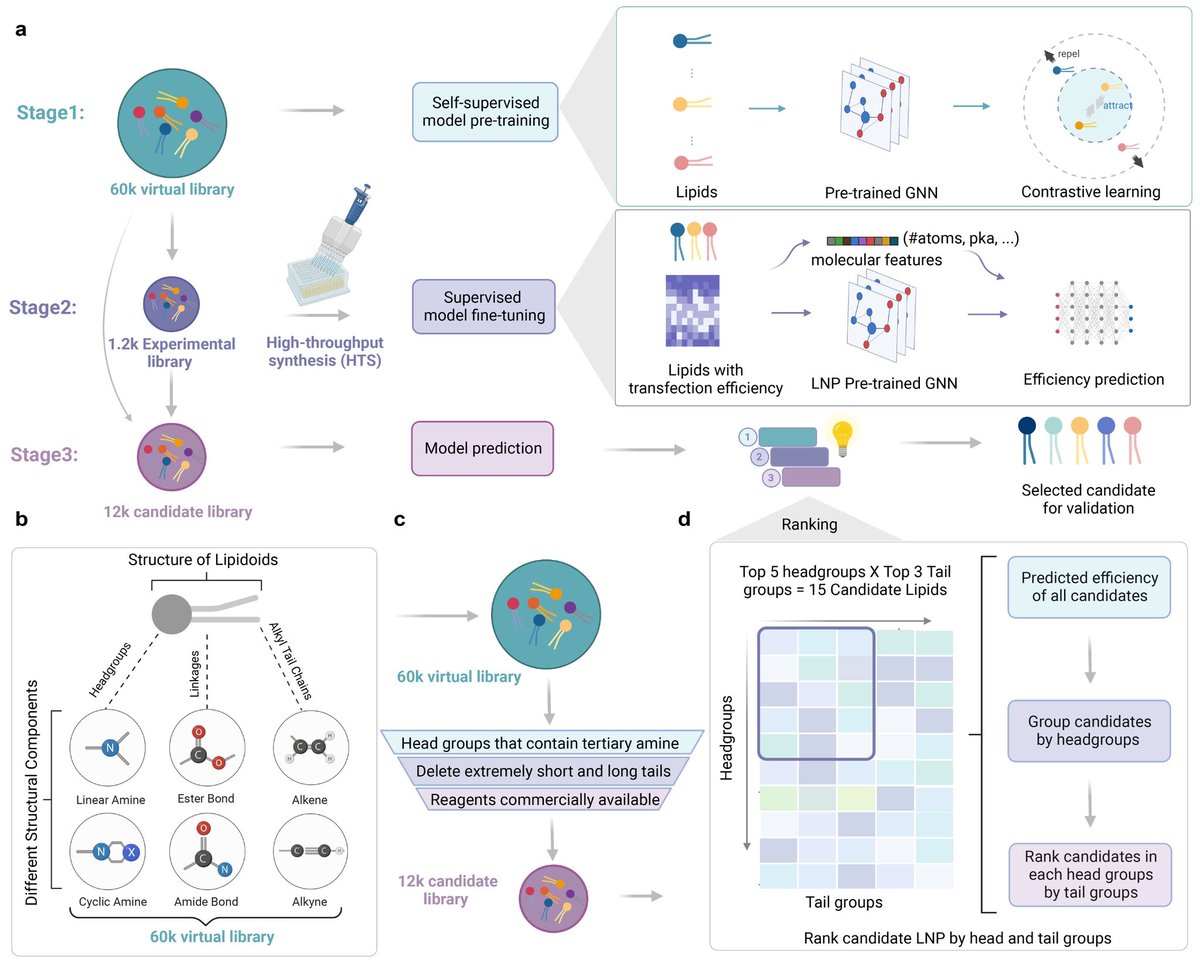
Thrilled to announce that our AGILE framework, an AI-guided LNP development for mRNA delivery, is now published in Nature Communications! 🎉 This is a collaborative work with @BowenLi_lab. (Re-)Introducing the AGILE Platform: A Deep Learning-Powered Approach to Accelerate LNP Development


Big congrats to Phil Fradkin , and such a great fitting name Orthrus!🎉


Thanks a lot! Eric Topol We discussed unique opportunities and a few promising techniques for building virtual cell models. I am also personally so exited about this direction. nature.com/articles/s4158…

In the third paper, Bo Wang Toronto General Hospital Research Institute & team unveil a powerful #AI model that learns from layers of biological data to decode how life works—poised to revolutionize #biotech. Stay tuned for a feature story… Read here➡️ nature.com/articles/s4158… (4/4) Haotian Cui