
Hannah Wayment-Steele
@hwaymentsteele
Biomolecular dynamics, big datasets, small boats. Soon: asst prof @UWBiochem. Currently: @JCCFund fellow with @DorotheeKern.
hkws.bsky.social
ID: 4437432767
https://waymentsteelelab.org 10-12-2015 12:27:22
537 Tweet
1,1K Followers
709 Following

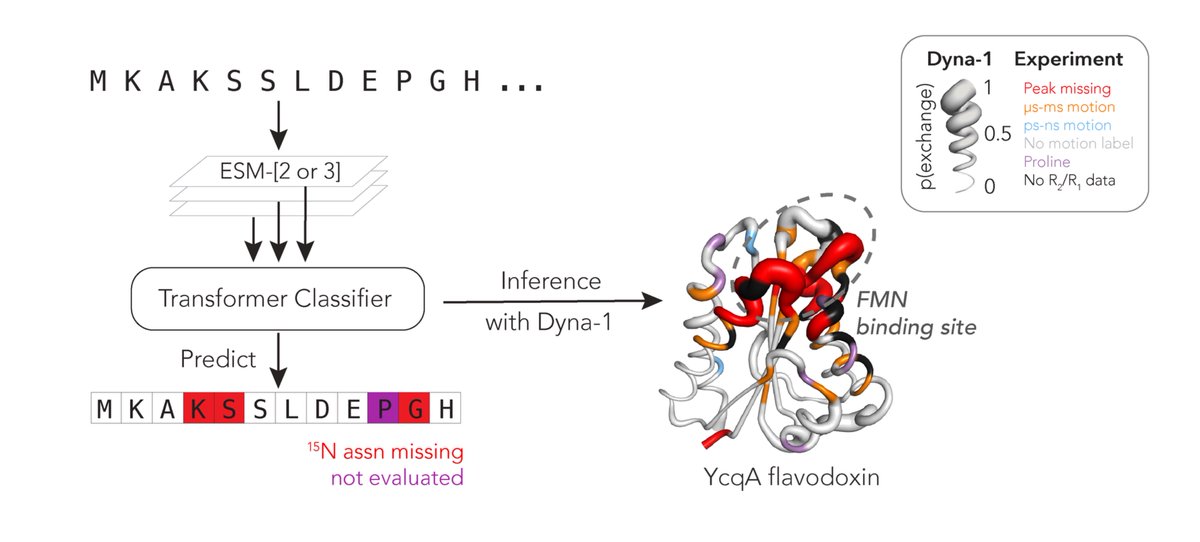
Protein function often depends on protein dynamics. To design proteins that function like natural ones, how do we predict their dynamics? Hannah Wayment-Steele and I are thrilled to share the first big, experimental datasets on protein dynamics and our new model: Dyna-1! 🧵

We collaborated with Hugging Face🤗to host all of our models and data online! Big thank you to Daniel van Strien Wauplin Quentin Lhoest 🤗 for helping out (and GPU)! Dyna-1 and our RelaxDB datasets on HF: 🚀huggingface.co/spaces/gelnesr… 🌟huggingface.co/datasets/gelne…


Egan Peltan This. Saying it louder for people in the back: *experiments* 🧪 bottleneck how fast AI can accelerate biology.
Next Tues (4/29) at **4:30PM** ET, we will have Gina El Nesr Hannah Wayment-Steele present "Learning millisecond protein dynamics from what is missing in NMR spectra" Paper: biorxiv.org/content/10.110… Sign up on our website for zoom links!

What could Alphafold 4 look like? (Sergey Ovchinnikov, Ep #3) 2 hours listening time (links below) To those in the (machine-learning for protein design) space, Dr. Sergey Ovchinnikov (Sergey Ovchinnikov) is a very, very well-recognized name. A recent MIT professor (circa early

Our team at Arcadia Science took a recent preprint on attention mechanisms for genotype–phenotype mapping, validated the results, improved the performance, and turned it into shareable research in just 9 days. 1/6 🧵 arcadia-science.github.io/2025-geno-phen…

DSI is hiring AI engineers, MLOps specialists, and a data scientist to support faculty research, with an emphasis on the UW–Madison RISE initiative. This is a great opportunity to participate in cutting-edge research across a wide range of topics. dsi.wisc.edu/about/work-at-…



for paper refs about this, I would refer everyone to start with doi.org/10.1016/j.pnmr… from Wim Vranken "In NMR, ensembles of structural models are calculated, with each model typically assigned a score or energy that relates how well they correspond to the generic modelling


