Hadeer Elhabashy
@hadeerelhabashy
Postdoc & Ph.D @MPI_BIO & @uni_tue | M.Sc in Physics @AUC | B.Sc in Biophysics @CairoUniv #LINO70 #LINO23
Structural Bioinformatics
ID: 246386776
02-02-2011 18:05:02
3,3K Tweet
1,1K Followers
3,3K Following


Today we report Evo, a generalist foundation model for biology, on the cover of Science Magazine. Trained on billions of DNA nucleotides, Evo was designed to capture two key aspects of biology: the multi-modality of the central dogma and the multi-scale nature of evolution.


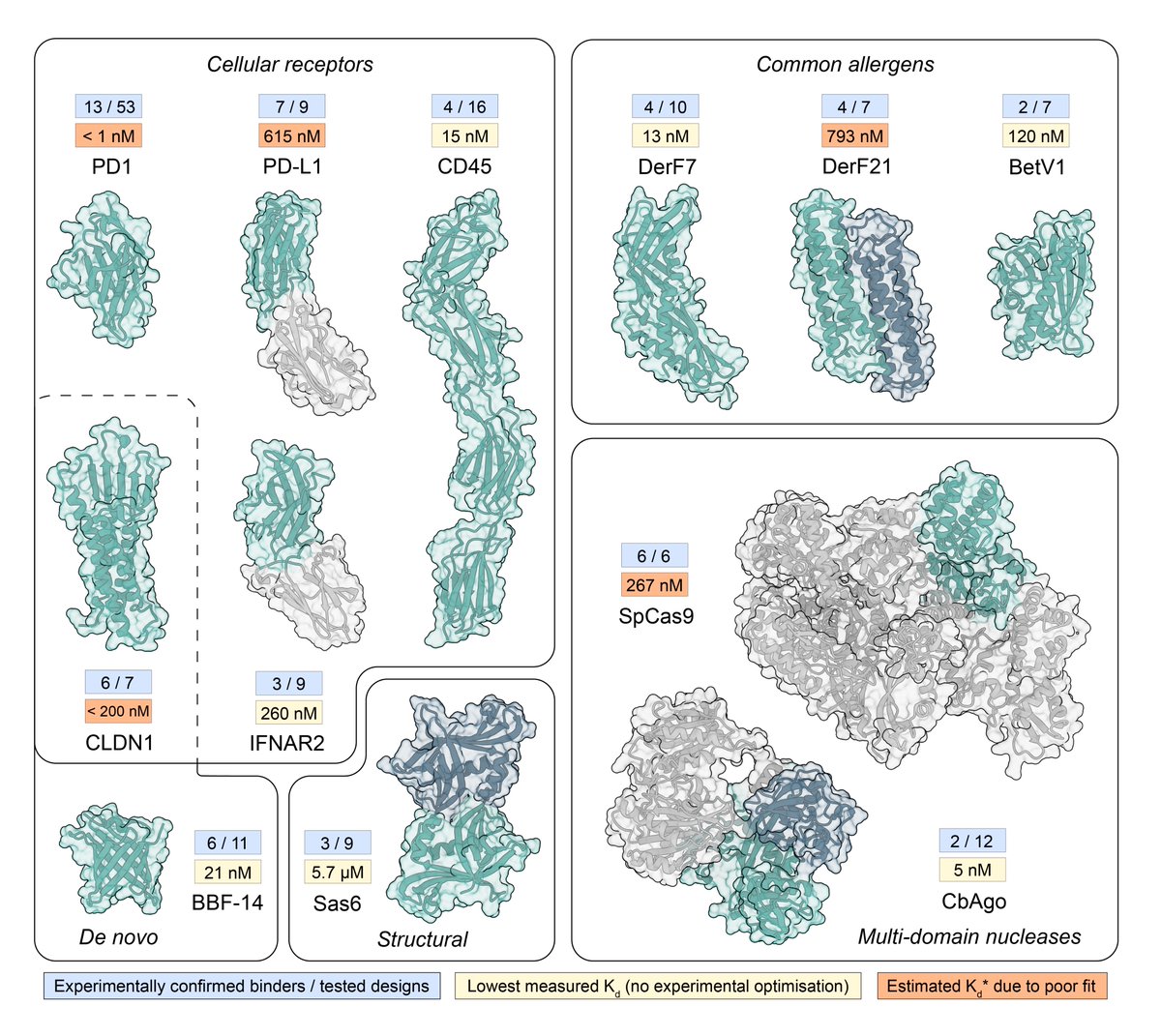
A few more letters... and we can finally write amino acid sequence with protein structure encoded by amino acid sequence? 🧬🤔😀 Christopher Frank Dominik Schiwietz Lara Fuss @hendrik_dietz biorxiv.org/content/10.110…








Starting 🔜 the The Nobel Prize lectures featuring our laureates - CEO @DemisHassabis and Research Director John Jumper - who will discuss their award-winning work on protein prediction and the journey to building #AlphaFold. Tune in to watch from 9:50am GMT →



It was a great morning today watching John Jumper, Research Director Google DeepMind, deliver his The Nobel Prize lecture on protein prediction and the journey to building #AlphaFold







Vacancy Alert ! ! Interested in #genomics, #bioinformatics and #AMR and working in an interdisciplinary team based at University of Oxford? We have a PostDoc position open now in our lab ‼️ Apply at ineosoxford.ox.ac.uk/careers Please RT and share !