
Henrik Dahl Pinholt
@henrikpinholt
PhD student at MIT Physics. Interested in statistics, active matter, single molecule biophysics and protein structure-function.
ID: 1324622024522801152
06-11-2020 07:58:07
37 Tweet
165 Followers
351 Following


Proteiners adfærd kan nu spores med kunstig intelligens 👉betyder at forskerne med stor nøjagtighed kan forudsige, hvad proteinet er ’god’ eller ’slem’ til. Fx proteiner, der får kræft til at opstå. Mere om det nye IA-værktøj bit.ly/3zVEDFT #dkforsk 📸Københavns Uni


Check out this news article about the work of Søren S.-R. Bohr, Josephine Iversen and I on taking fingerprints of diffusing proteins with machine learning. Shoutout to the lab of Nikos Hatzakis lab and Wouter Boomsma for making the study a reality! sciencenews.dk/en/identifying…


Loops in chromosomes leave a distinct signature in Hi-C data. A new polymer model - a chain folded into loops - can be solved analytically, revealing otherwise invisible loops. Great teamwork: Hugo Brandao, Kirill Polovnikov Sergei Belan, Max Imakaev.


Did you know that it’s possible to map out an entire reaction rate network by single molecule #microscopy? Too see it done for insulin heximerization, check out our latest preprint by Freja Bohr from Nikos Hatzakis lab and Knud J. Jensen labs. Amazing work! 🔬🔬 shorturl.at/ehixN


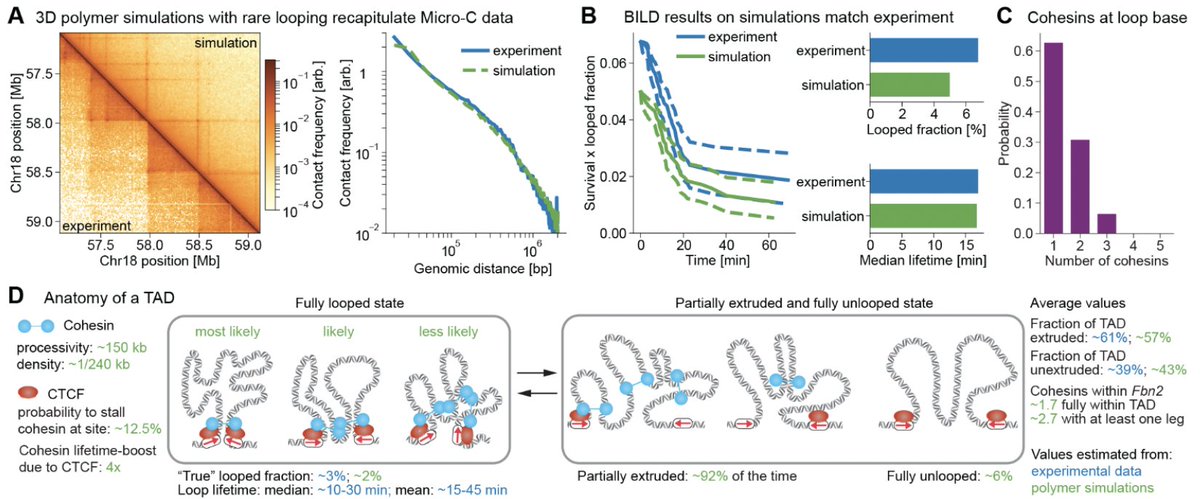
(1/6) our preprint biorxiv.org/content/10.110… is now out in Science Magazine science.org/doi/10.1126/sc… Using super-res live-imaging we show that CTCF/cohesin loops are dynamic (~10-30 min median lifetime), rare (~3-6.5% looped fraction), and we fully parameterize a TAD.


We had such a fantastic time with my group (and 3 alumni) in Department of Chemistry, University of Copenhagen celebrating this year's papers in Nature Chemistry and Nature Communications with a BBQ and cocktails. I am so proud and thankful for each one of you !



Super excited to see our recent work out in Communications Biology .We developed a super resolution method #REPLOM for real time observation of heterogeneous protein aggregation and extraction of heterogenous growth kinetics. Great collaboration with FoderàLab nature.com/articles/s4200…

Kudos to Min Zhang, for relentlessly driving this through over 2 years of intense work, and also Xin Zhou, Henrik Dahl Pinholt, Søren Bak, Luca Banetta and Alessio Zaccone 🇺🇦 . work was done at Department of Chemistry, University of Copenhagen and Funding from Lundbeckfonden and Villum Fonden Science at VILLUM

(1/n) How do active processes affect chromatin folding, and how does this compare to passive, affinity-driven polymer folding? New preprint out 👉biorxiv.org/content/10.110… with Andriy Goychuk, Arup K. Chakraborty, and Mehran Kardar and first paper of my PhD MIT Physics 🧬


It took Sergey Ulyanov and I a while to shape fountains, a novel type of chromatin architecture. Here is our story of pioneering factors binding (Daria Onichtchouk) and the loop extrusion (Leonid Mirny) working together. (can you tell that I’ve been into fishkeeping since I was six?)



1/ My work on epigenetic memory with Leonid Mirny, Dino Osmanovic is out Science Magazine! science.org/doi/10.1126/sc… How can *stable* memory be encoded in patterns of *dynamic* epigenetic marks? We find 3D folding of genome can stabilize memory, if 3 ingredients are present...
![Alex Cagan (@atjcagan) on Twitter photo Incredibly excited to share our paper ‘Somatic mutation rates scale with lifespan across mammals’ now published @nature.
nature.com/articles/s4158…
An illustrated & updated tweetorial… [1/24] Incredibly excited to share our paper ‘Somatic mutation rates scale with lifespan across mammals’ now published @nature.
nature.com/articles/s4158…
An illustrated & updated tweetorial… [1/24]](https://pbs.twimg.com/media/FQPMMCCWYAIybBt.jpg)