Jay Shendure
@jshendure
Genomics technology developer. Developmental biology adult learner. Professor at @uwgenome @HHMINEWS @BrotmanBaty. He/him.
ID: 296644509
https://krishna.gs.washington.edu/ 11-05-2011 04:46:54
508 Tweet
13,13K Followers
318 Following

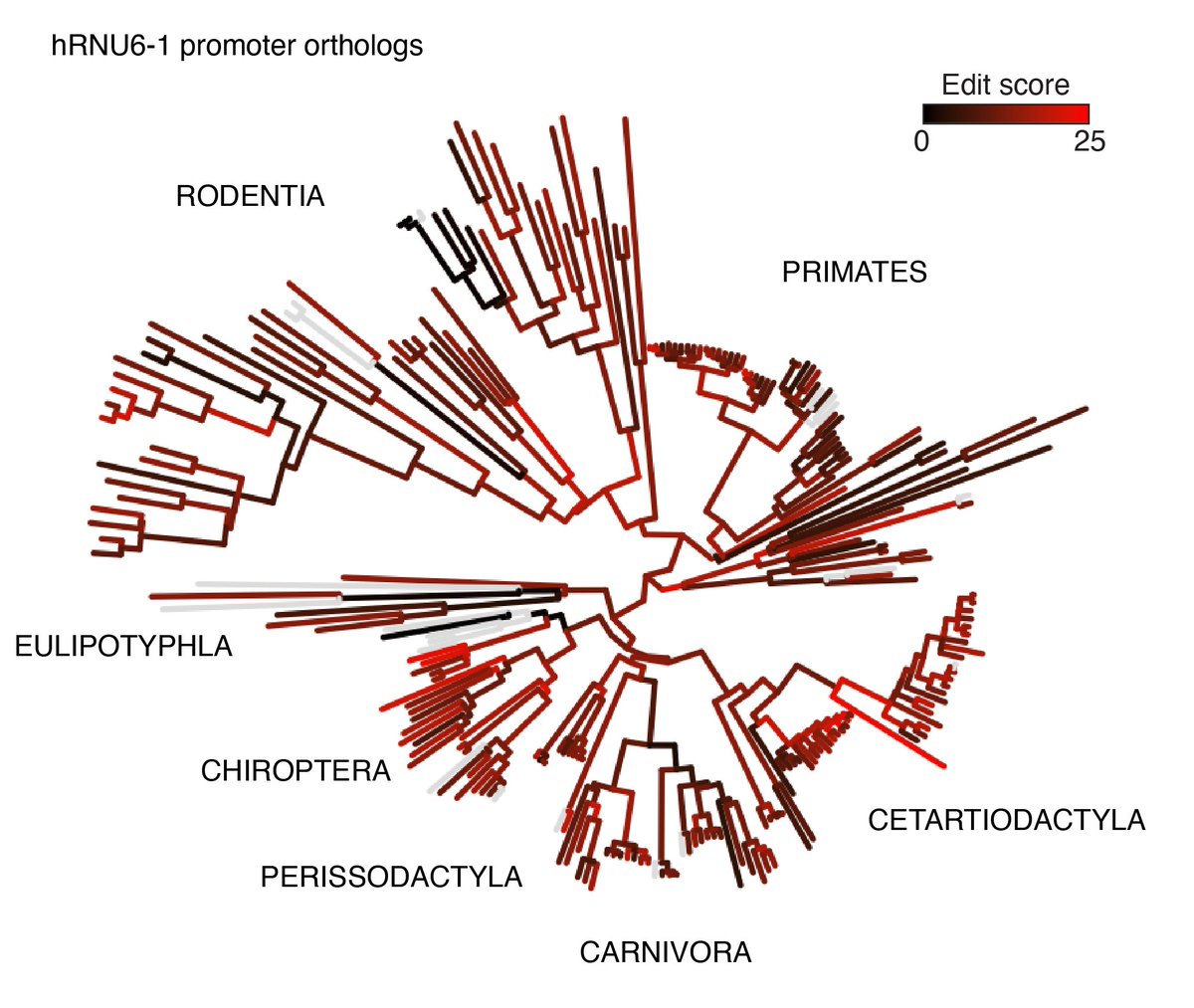
Excited to share my postdoc work from the Alex Schier. This project was a joint effort with many collaborators Georg Seelig, Sebastián Castillo-Hair,Lucia Du, Yiqun Wang,Adam Carte,Mariona Colomer Rosell, Chris Yin. 1/20 biorxiv.org/content/10.110…

Deluge of useful information @ proteomics, protein/mRNA correlations, comparative molecular landscapes and more. #gastruloids (#mouse and #human) biorxiv.org/content/10.110… Valuable resource. Huge effort from Devin Schweppe Nobu Hamazaki and colleagues #EmbryoModels #InNumbersWeTrust


Very happy to share our latest w/ Devin Schweppe and Nobu Hamazaki labs, where we map the temporal dynamics and proteomic landscape of mammalian gastruloids. Sneek Peak 🧵below #stembryo #gastruloid #proteomics biorxiv.org/content/10.110…

💫NEW: Hamazaki, Yang et al. report that an early pulse of #RetinoicAcid induces human #gastruloids with a neural tube, segmented somites and more advanced cell types than conventional gastruloids. #embryo_models wei yang Nobu Hamazaki Jay Shendure nature.com/articles/s4155…





Incredibly proud to share that I recently joined UCSF Bioengineering & Therapeutic Sciences UC San Francisco as an Assistant Professor! This means that the Calderon Lab is now open! Or, more accurately, in the process of opening as I am still unboxing equipment and unraveling giant posters (a last @JSHendure lab gift) 🥳





📢 Check out our teamstembryo Nature Cell Biology News & Views with postdoc Alexandra Schauer on the latest feat in human stem-cell-based embryo models by Nobu Hamazaki, Jay Shendure & colleagues. Full text: rdcu.be/dWw3c nature.com/articles/s4155… #gastruloids





Beyond thrilled to present our O-MAP—now *published* Nature Methods (!!!) O-MAP is a powerful new method for biochemically "dissecting" the subcellular microenvironment around an RNA of interest, using off-the-shelf parts and standard manipulations. 🧵 nature.com/articles/s4159…