
Jiacheng Miao
@jiacheng_miao
Biomedical Data Science Grad Student @UWMadison // StatGen & AI/ML
ID: 1066568847073923073
https://jiachengmiao.com 25-11-2018 05:46:43
221 Tweet
399 Followers
1,1K Following



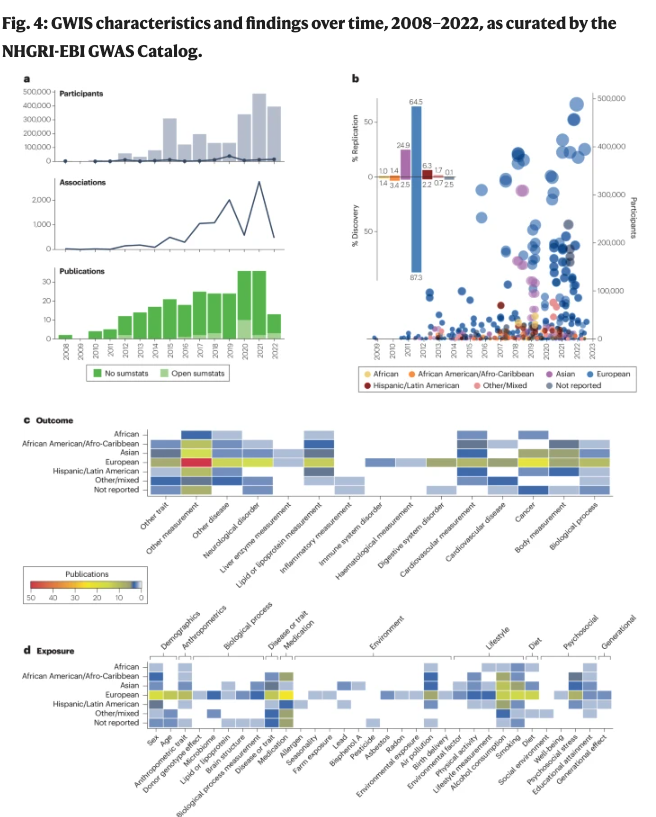
Interested in GxE interaction #gwas? Now, you can identify and filter them on our website! Check the great review made by Genevieve Wojcik, PhD MHS et al. at @JohnsHopkinsEPIThey. They also analysed the data we hosted and showed a lack of #SumStats available. tinyurl.com/bdfmsmtd


I and my colleague, Sahar Gelfman, at RGC had the privilege to write a News and Views article for @nature about a recent work on Huntington's disease by Steve McCarroll and team. In this incredible work, through an innovative single cell RNA sequencing methodology, Handsaker et




Story by Alvin Powell on Sonia Vallabh and Eric Minikel’s very personal mission with our lab and others to develop a gene editing treatment for prion disease: news.harvard.edu/gazette/story/…. The research paper in Nature Medicine can be found below. prionalliance




In this Article, the authors introduce PIGEON, a statistical framework for estimating gene-environment interactions for complex traits. Jiacheng Miao Qiongshi Lu nature.com/articles/s4156…

Jiacheng Miao's PIGEON paper on GxE methodology is now published Nature Human Behaviour with a new title. In our view, this paper can reshape the study design for future complex trait GxE work. Paper📰doi.org/10.1038/s41562… Software🧑💻github.com/qlu-lab/GSUB

Excited to share our latest preprint on the impact of background selection on complex trait evolution. It’s amazing to finally see one of the major projects from my PhD journey come to life. Huge thanks to Jeremy J. Berg and John Novembre for their guidance and support.

I’m very excited to be joining MIT Biological Engineering MIT Dept of BE as an Assistant Professor and the Ragon Institute Ragon Institute as a member in January 2026! I will be recruiting students and postdocs (see more info below).














