Julia Rogers, PhD
@juliarurogers
Computational+biophysical chemist venturing into systems biology+ML as a @JCChildsFund postdoc @Columbia | PhD @UCBerkeley | BS @TuftsUniversity | she/her
ID: 1133068894455836672
27-05-2019 17:54:10
260 Tweet
799 Followers
1,1K Following



Super excited to preprint our work on developing a Biomolecular Emulator (BioEmu): Scalable emulation of protein equilibrium ensembles with generative deep learning from Microsoft Research AI for Science. #ML #AI #NeuralNetworks #Biology #AI4Science biorxiv.org/content/10.110…

🚨NEW PUBLICATION ALERT🚨: In a paper out today in Science Magazine we describe a way to build synthetic phosphorylation circuits with customizable sense-and-response functions in human cells. Check it out at science.org/doi/10.1126/sc…. 🧵1/n




Structural biology is in an era of dynamics & assemblies but turning raw experimental data into atomic models at scale remains challenging. Minhuan Li and I present ROCKET🚀: an AlphaFold augmentation that integrates crystallographic and cryoEM/ET data with room for more! 1/14.

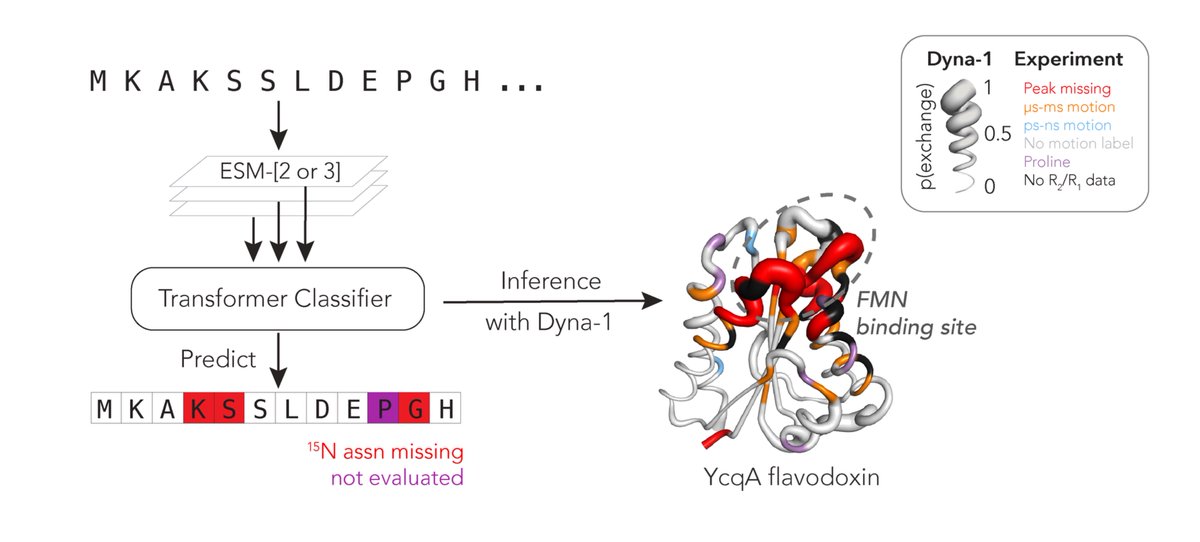
Protein function often depends on protein dynamics. To design proteins that function like natural ones, how do we predict their dynamics? Hannah Wayment-Steele and I are thrilled to share the first big, experimental datasets on protein dynamics and our new model: Dyna-1! 🧵



Awesome to see this epic piece of work, led by Cecilia Clementi, finally appear in Nature Chemistry. The development of a general coarse-grained protein forcefield to describe folding, binding and conformation changes without solvent and all-atom, has been long anticipated!


The MLSB workshop will be in San Diego, CA (co-located with NeurIPS) this year for its 6th edition in December 🧬🔬 Stay tuned MLSB (in San Diego) as we share details about the stellar lineup of speakers, the official call for papers, and other announcements!🌟


Excited to share work with Zhidian Zhang, Milot Mirdita, Martin Steinegger, and Sergey Ovchinnikov biorxiv.org/content/10.110… TLDR: We introduce MSA Pairformer, a 111M parameter protein language model that challenges the scaling paradigm in self-supervised protein language modeling 🧵