
Karen Miga
@khmiga
Associate Professor at UCSC; UC Santa Cruz Genomics Institute; Satellite DNA Biologist
ID: 2344030220
https://www.migalab.com/ 14-02-2014 19:41:49
1,1K Tweet
3,3K Followers
1,1K Following

🧬Want to learn more about the Telomere-to-Telomere project and the folks involved in the T2T Consortium before the T2T-F2F meeting on Sept 3-4? 👇Watch the presentation Karen Miga gave at the Genome Technology Forum held The Jackson Laboratory in collab w/ National Human Genome Research Institute youtu.be/t4kGX373EEQ

Our fast haplotagging paper is now published (nature.com/articles/s4146…) - see the earlier thread for a summary. Thanks to Kishwar Alexey Kolesnikov for leading the work and collaborators: @Johngorzynski, Sneha D. Goenka, euan ashley, Miten Jain, Karen Miga, @BenedictPaten



Genome sequencing is faster and cheaper than ever – but obstacles to broad adoption remain. Join Google’s genomics lead Andrew Carroll & UC Santa Cruz Genomics Institute’ @BenedictPaten for a live-streamed discussion on genomic analysis and equity: goo.gle/3ZmPyYO


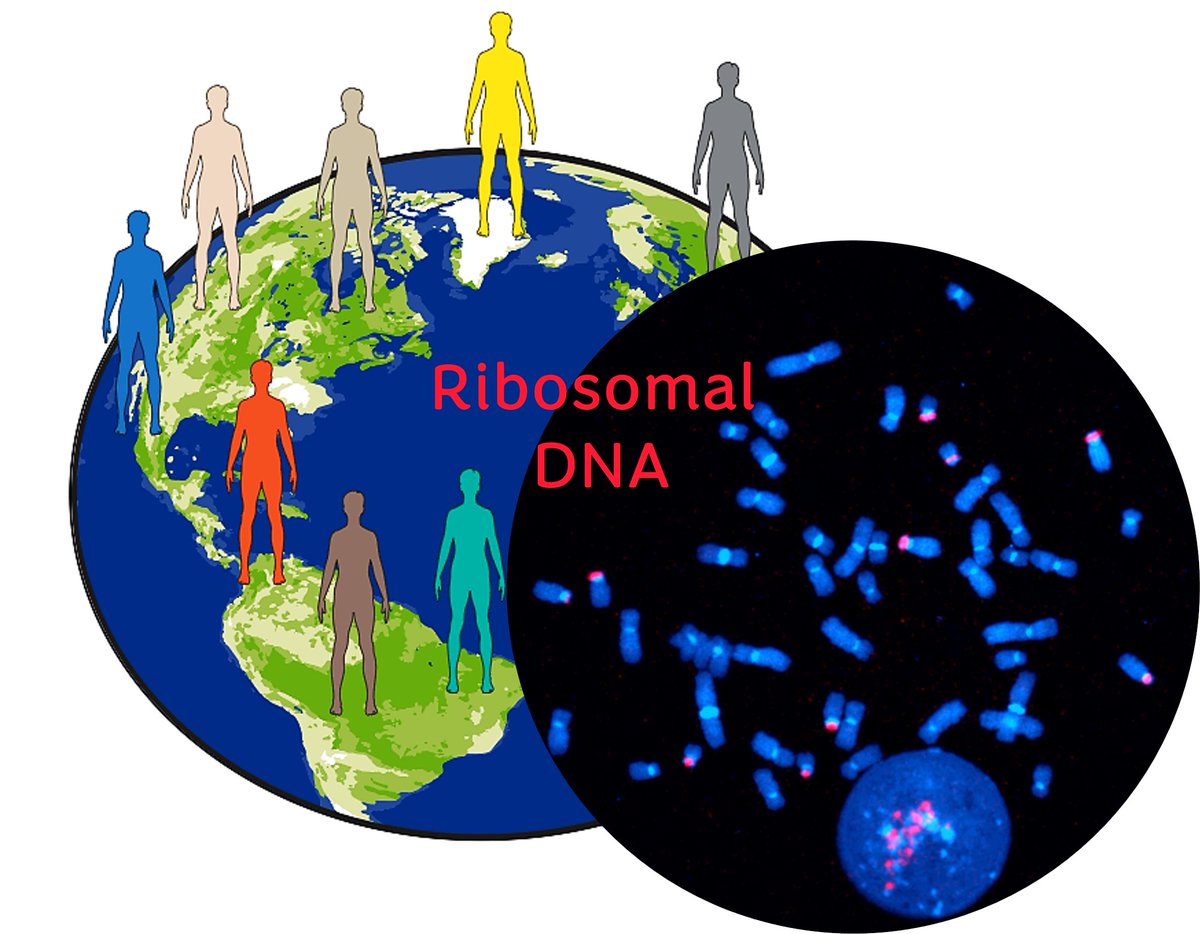
New preprint from our lab 🚨 We explored the variability in ribosomal DNA copy number and activity across humans of the world with Jennifer Gerton Paxton Kostos Andrea Guarracino Steven Solar Erik Garrison Adam Phillippy and others not on X biorxiv.org/content/10.110… 🧵



Accurate genome assemblies are key for research. DeepPolisher uses Pacbio HiFi alignments to improve base-level accuracy, cutting errors by half and reducing indel errors by over 70%. ✍🏼Mira Mastoras Kishwar @BenedictPaten Mobin Asri Here's how... biorxiv.org/content/10.110…






Very excited to share our work Megan Mahlke in great collaboration with Nicolas Altemose Karen Miga Peter Campbell and Rachel O'Neill revealing that human centromeres are heterogenous and naturally evolve over prolonged proliferation. Check it out here biorxiv.org/content/10.110…








