Elodie Laine
@laineelodie
Full Professor @Sorbonne_Univ_, working @LCQB_UMR7238 on protein evolution, structures, and interactions. @laineelodie.bsky.social
ID: 1255513629673357322
http://www.lcqb.upmc.fr/laine 29-04-2020 15:05:56
536 Tweet
521 Followers
375 Following


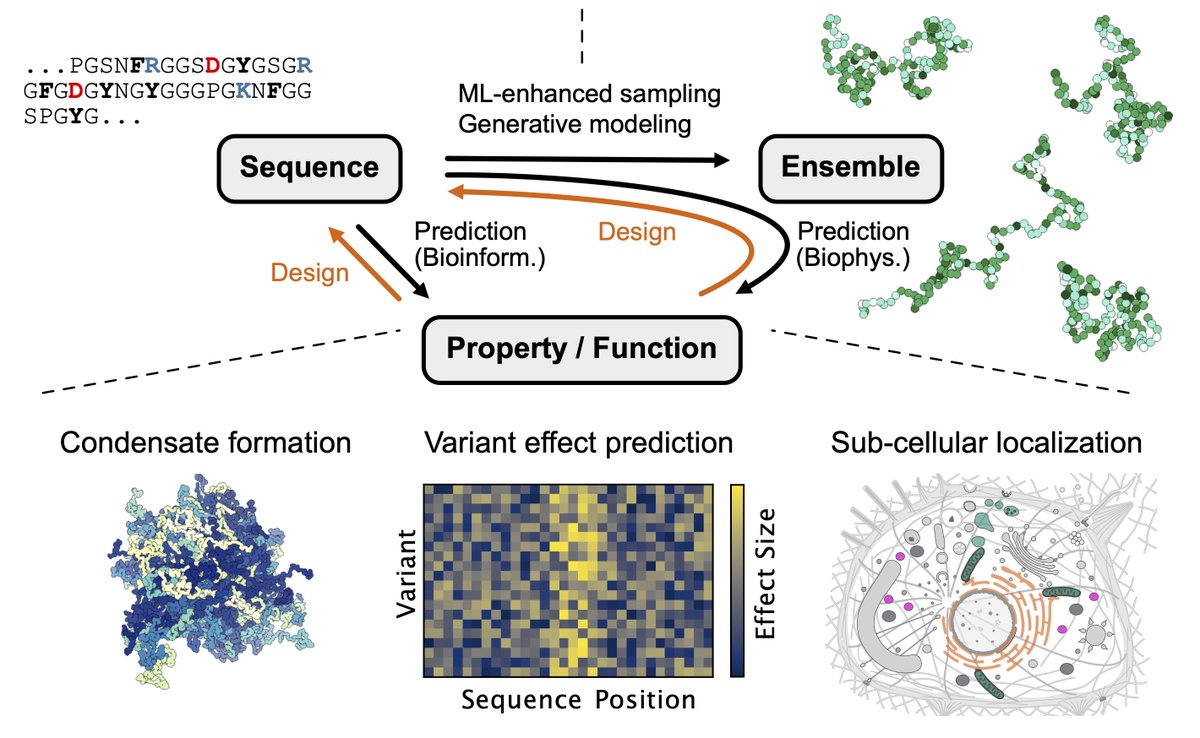
We are pleased to announce the release of The Protein Design Archive. Our comprehensive database of designed proteins highlights the triumphs and challenges of the field: pragmaticproteindesign.bio.ed.ac.uk/pda/ Preprint:biorxiv.org/content/10.110… Michael Stam Marta Chronowska WoolfsonLab #proteindesign

As part of my recently funded ANR-JCJC grant ANR - Agence nationale de la recherche, I’m looking for a postdoc researcher and an engineer with background in deep learning for protein science. Please contact me if you’re interested. Centre Inria de l'Université de Lorraine Loria

I’m thrilled to announce: at JURA Bio, Inc. we’ve constructed a new kind of generative protein model, which allows us to manufacture its generated designs in the real world at petascale. Paper: static1.squarespace.com/static/66db3a5… Blog: jura.bio/blog/variation…







Happy to share our latest on using Alphafold2 with experimental distance restraints to model protein conformations. Vanderbilt School of Medicine Basic Sciences Vanderbilt Center for Structural Biology Google DeepMind #alphafold biorxiv.org/content/10.110…






Curious about alternative splicing? Kick off the year with our latest review in Current Opinion/Research in Structural Biology! authors.elsevier.com/sd/article/S09…