
Lars Borm
@larsborm
Postdoc at Stein Aerts lab, smFISH of the non-swimming kind. Building and tinkering.
ID: 919866187164635136
16-10-2017 10:02:44
348 Tweet
566 Followers
219 Following

So excited to announce our new method for multiplexed RNA imaging in bacterial cells: bacterial-MERFISH! A huge congratulations to the team of Ari Sarfatis, Yuanyou Wang, and Nana Twumasi-Ankrah! Check out the following thread or our bioRxiv (biorxiv.org/content/10.110…) 1/12


A great DIY guide for image-based spatial transcriptomic from Florian Mueller and team at Institut Pasteur, since 1887 biorxiv.org/content/10.110…




🚨𝐇𝐞 𝐋𝐚𝐛 job opportunity UC San Francisco 🚨: I'm hiring TWO postdocs - an experimentalist and a computational biologist. Join us to use cutting-edge single-cell and spatial technologies to unravel the mysteries of human tissues. opportunities.ucsf.edu/content/open-p…
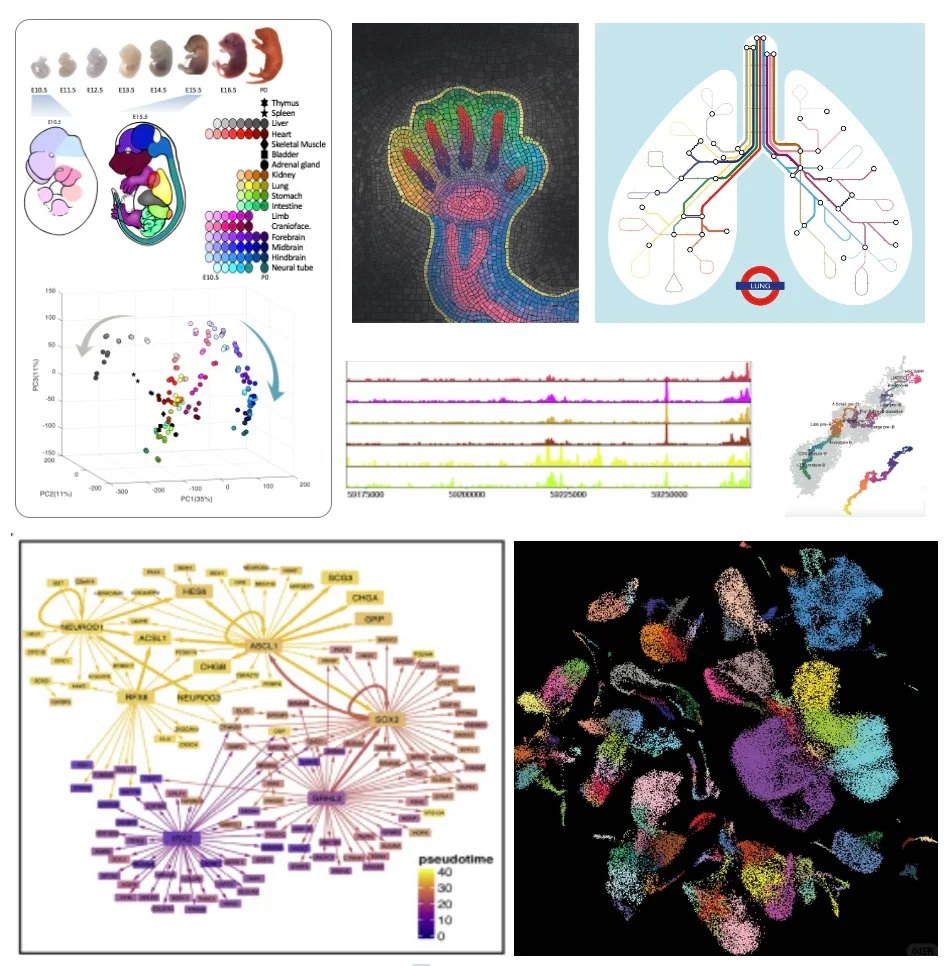


🎉 Exciting news! Our latest work is now published in Development! The open-source solution for RNA-targeted in situ sequencing provides an accessible and efficient method for spatially resolved gene expression analysis and spatial multi-omics. 🔗 doi.org/10.1242/dev.20…

Interested in navigating #metabolism in space and time? Meet uMAIA (the Unified Mass Imaging Analyzer) - a framework developed in collaboration with Giovanni D'Angelo & oaties EPFL Life Sciences led by Halima Hannah Schede & Leila #omics #lipidtime #MALDI biorxiv.org/content/10.110… 1/N


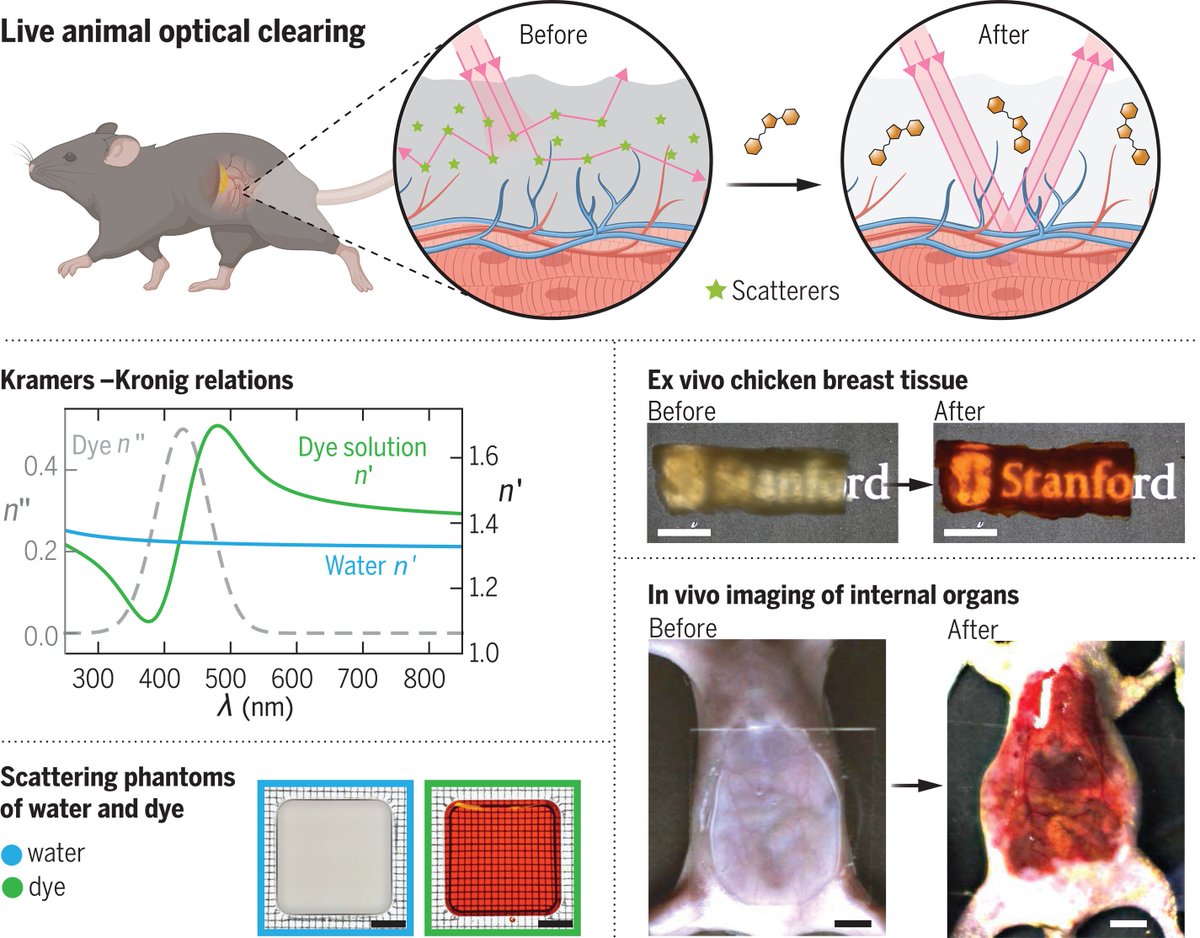
Long awaited (thanks James Manton for posting the patent application!) and now out in Science Magazine: Using absorption to look deeper by HongLab - they achieved approx. 3 mm depth using the food dye tartrazine: #Microscopy science.org/doi/10.1126/sc…

7 years ago, I met a junior fellow named Jason Buenrostro who blew me away with a vision of futuristic genomic technologies Today, we (Ajay Labade, @carolinecomenho) are excited to share our first steps into that future: Expansion in situ genome sequencing 1/

our work on vitessce is out in Nature Methods!


STAMP: Single-Cell Transcriptomics Analysis and Multimodal Profiling through Imaging | bioRxiv Dr. Jasmine Plummer 👩🏻🔬🧬🇨🇦🇺🇸 Holger Heyn @pascual_reguant @eliseinsing CNAG Adelaide Centre for Epigenetics St. Jude Research biorxiv.org/content/10.110…





