Lucía Barbadilla
@luciabarmar
PhD student @LabRidder | Studying noncoding variants & gene regulation with Deep Learning
ID: 1232327458902417409
25-02-2020 15:32:32
25 Tweet
56 Followers
262 Following


In our latest episode of Roman Cheplyaka | the bioinformatics chat, we talk with Žiga Avsec about research in academia vs industry, Enformer, and deep learning libraries! Great to hear about the work directly from the source. Hope other people enjoy our conversation! bioinformatics.chat/enformer


#ScienceForUkraine If you are a postdoc, PhD or Master's student interested in Cancer Genomics and would like to join our lab (bbglab) for an internship contact me. List of labs supporting Ukrainian Scientists. docs.google.com/spreadsheets/d… 🇺🇦#ScienceForUkraine #ScienceForUkraine 🇺🇦





‼️ ONLINE NOW Nature Genetics 📰 A sequence-based global map of regulatory activity for deciphering human genetics 🧑🏿🤝🧑🏻 Jian Zhou Olga Troyanskaya and team 👇🏻 go.nature.com/3ACPk3m



Impactful words by Carolyn Bertozzi today on how diversity in her trainees enabled creativity, not playing by the rules, and ultimately the work that built the field of bioorthogonal chemistry. Diverse science leads to better science!

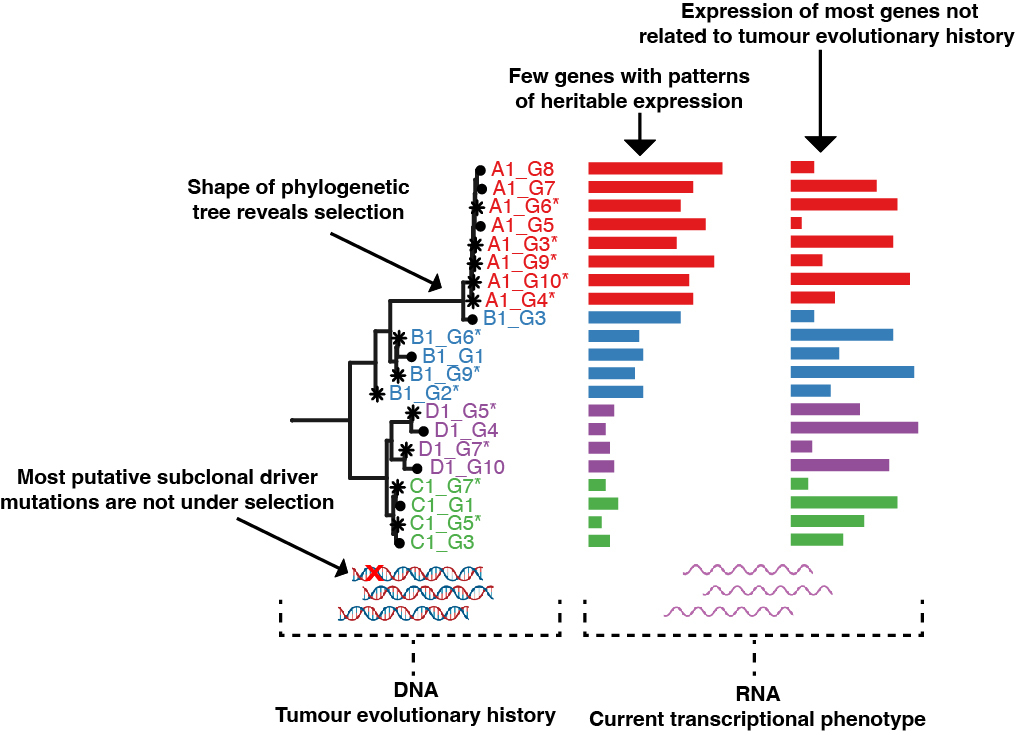
How do the #genome, #epigenome and #transcriptome co-evolve in colorectal #cancer? In the second of two papers in @nature, we find gene expr is not strongly heritable & evaluate "driverness" of muts directly in patient tumours nature.com/articles/s4158… Andrea Sottoriva #EPICC2 1/7