MINT database
@mint_database
MINT is a public database developed at the University of Tor Vergata in Rome and it focuses on experimentally verified protein-protein interactions.
ID: 885437741022019584
http://mint.bio.uniroma2.it/ 13-07-2017 09:56:22
241 Tweet
294 Followers
313 Following

Can't recommend this free 2-day #GeneOntology workshop enough!! If you can get to Rome the week after #biocuration2023, sign up now. Space is limited and yes the instructors are absolutely fantastic! Functional Gene Annotation MINT database

Kicking off the main track of the meeting, Ruth Lovering (Ruth Lovering) is giving a greeting from Int Soc Biocuration society #biocuration2023


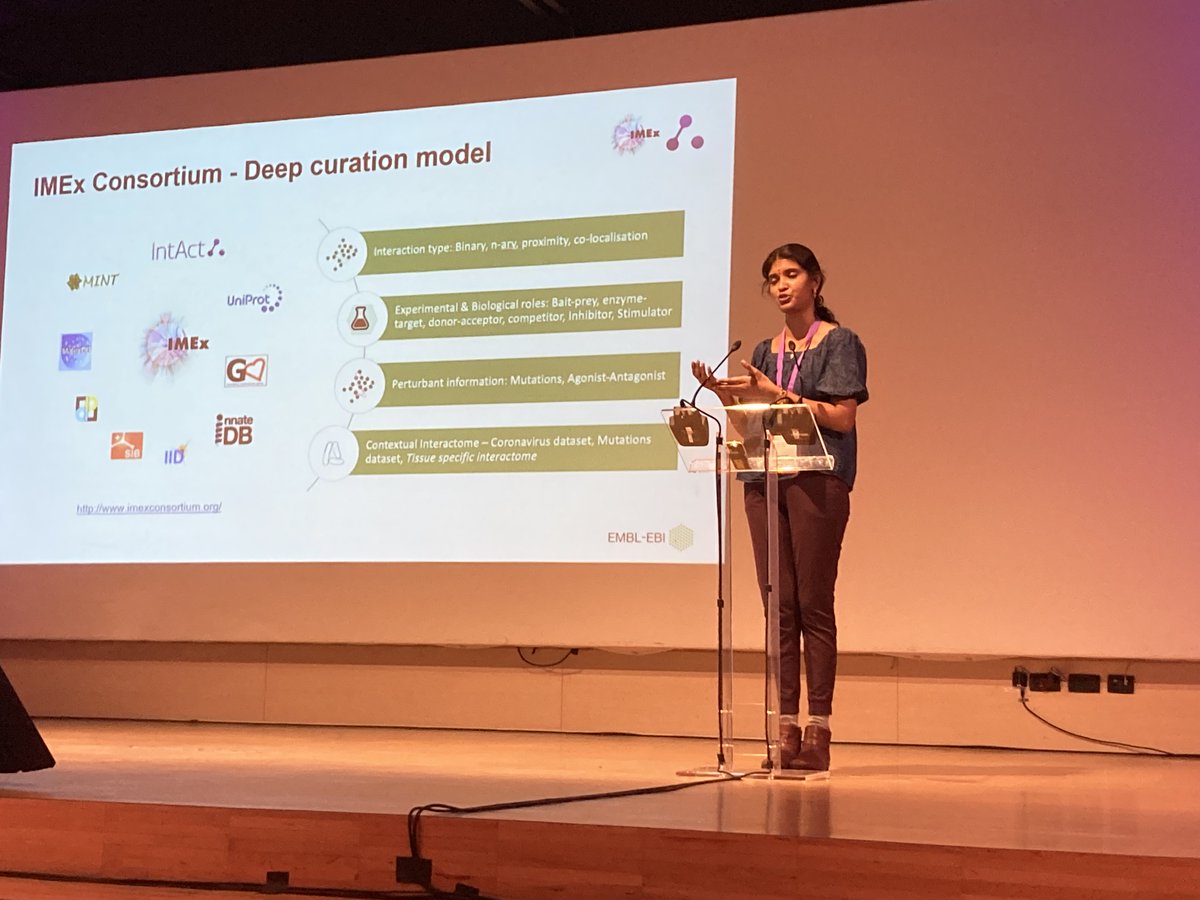
Great talk of Kalpana Panneerselvam from IntAct at EBI team on imexconsortium’s contextual metadata and interactomes at the Biocuration 2023 day 2. #biocuration2023





Emidio Capriotti highlighting the connection between Rare Diseases and Machine Learning ELIXIR Europe #ELIXIR23. Really interesting to see the overall assessment efforts around pathogenicity prediction - a lot of opportunities for #MachineLearning!



New interactions curated by MINT team are available via MINT database , IntAct at EBI and JG Kim website after the new IntAct release!!! x.com/intact_project…


Without biodata resources, there is no advance in the life and biomedical sciences. In its 2033 selection, GlobalBiodata has announced a further 15 Global Core Biodata Resources, bringing the set up to 52, collectively serving a breadth of domains, data types & user communities.

The GCBRs provide an essential data foundation for the life sciences, both directly to users and as data providers to more specialised topic-focused resources. GCBR leaders work with GlobalBiodata to explore, and drive towards, greater sustainability for biodata resources.

We are proud to announce we have been awarded Global Core data resource status by the @GlobalBiodata Coalition. Congratulations to all our member databases: IntAct at EBI, DIP, MINT database, MatrixDB, IID, InnateDB, UniProt, and also SIB. globalbiodata.org/global-biodata…



We are so pleased that imexconsortium has been included among the @GlobalBiodata resources! MINT database is a founding and an active member of IMEx. IMEx resources have been work jointly to share curation effort and provide a unique high quality protein interactions dataset

Our new paper "The landscape of cancer-rewired GPCR signaling axes" is out on Cell Genomics cell.com/cell-genomics/… Huge work by chakit arora and the bioinformatics group of Scuola Normale. Great collaboration with GutkindLab, Natoli IEO and other groups!


It's time to celebrate!!! imexconsortium