Marcin Grzybowski
@marcingrzy
ID: 1574705921044905984
27-09-2022 10:22:18
30 Tweet
80 Followers
106 Following

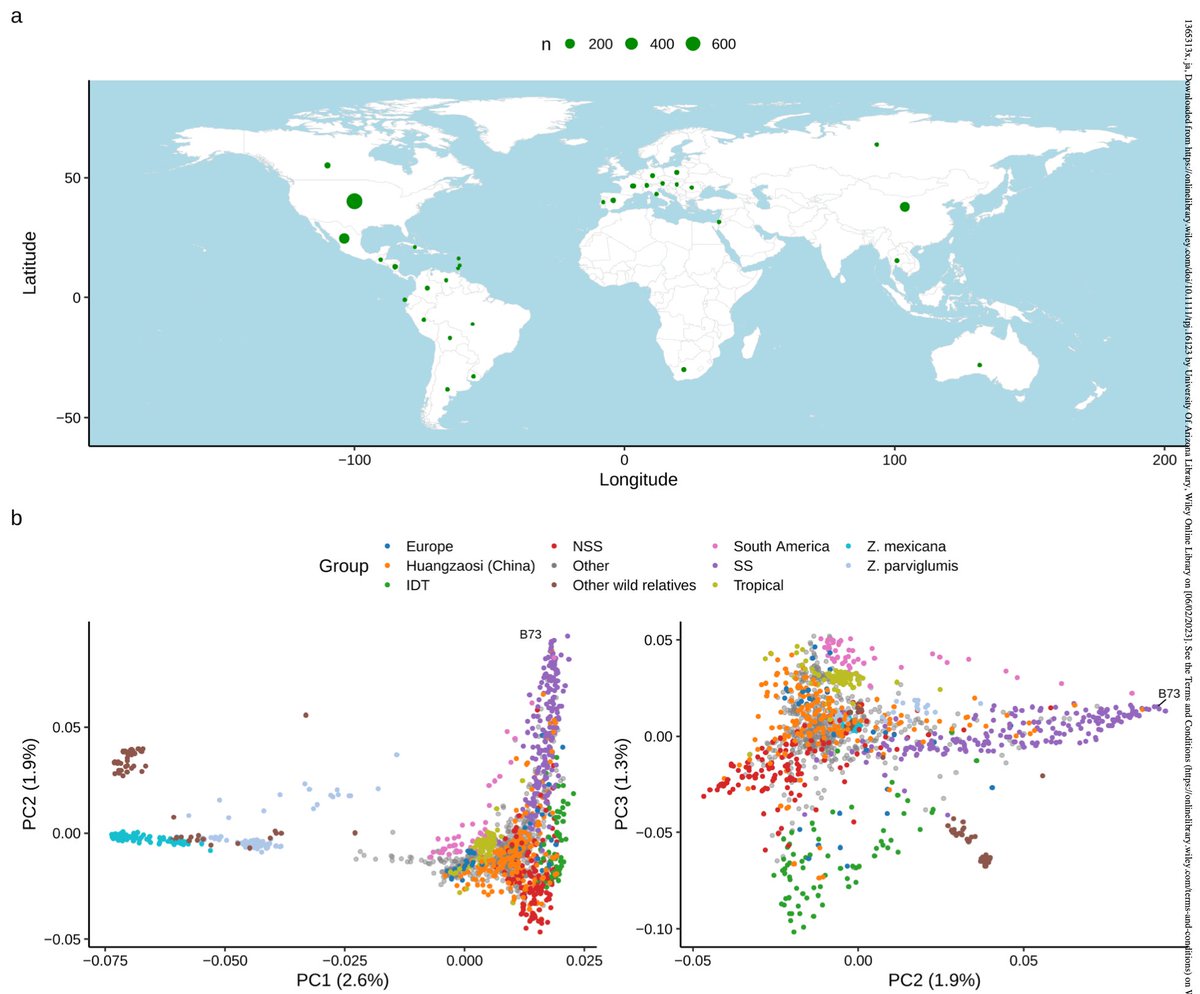
SEB invites you to read the March 2023 Editor Choice: "A Common Resequencing-Based Genetic Marker Dataset for Global Maize Diversity" showcased by the brilliant Gwendolyn Kirschner, from The Plant Journal🌱 Read now ⬇️ sebiology.org/resource/tpj-m… Happy reading




A natural surge protector buffers the delicate photosynthetic systems of corn against sudden changes in light intensity. Can we use existing genetic diversity to fine tune that surge protector? Congratulations Seema Sahay & Marcin Grzybowski! doi.org/10.1111/nph.18…


Genetic control of photoprotection and photosystem II operating efficiency in plants Sahay et al. Kasia Glowacka James Schnable UNL PSI Nebraska Biochem 📖ow.ly/AYxn50OuzMA

Kasia Glowacka Marcin Grzybowski Seema Sahay Scott Schrage | @scottschrage.bsky.social: You folks made page 3 of the print edition of the York News Times. Which may be one of the most ingenious names for a newspaper ever.





Genetically controlled differences between how photosynthesis changes in different corn plants stressed by a lack of nitrogen. New preprint from Seema Sahay @grzybowski Marcin Grzybowski & Kasia Glowacka. biorxiv.org/content/10.110…



Our resequencing for a common maize genetic marker dataset paper selected as a MaizeGDB Editorial Board pick! Specifically calls out how this enable "remixing and reuse" of genetic and phenotypic trait data. 😀Marcin Grzybowski The Plant Journal maizegdb.org/hot_new_papers…

In a new paper in Nature Plants we IDed a mechanism how the host plant genotype (🌽) contributes to the recruitment of its root microbiome. We identified a 🌽 gene that modulates lateral root density by regulating the abundance of specific bacteria under N-deficiency in soil. 1/2

Very impressed with my first experience publishing in Journal of Plant Physiology. Knowledgeable and thoughtful reviews and only 10 days from "it's accepted!" to the complete journal formatted PDF up online. Five stars. Would submit again. sciencedirect.com/science/articl…


Using autoencoders -- an unsupervised ML approach -- to derive plant traits from hyperspectral reflectance data. Potentially bypasses the need for extensive labeled datasets. Big thank you to lead author Michael Tross and our @aii4ra collaborators. doi.org/10.1002/ppj2.2…



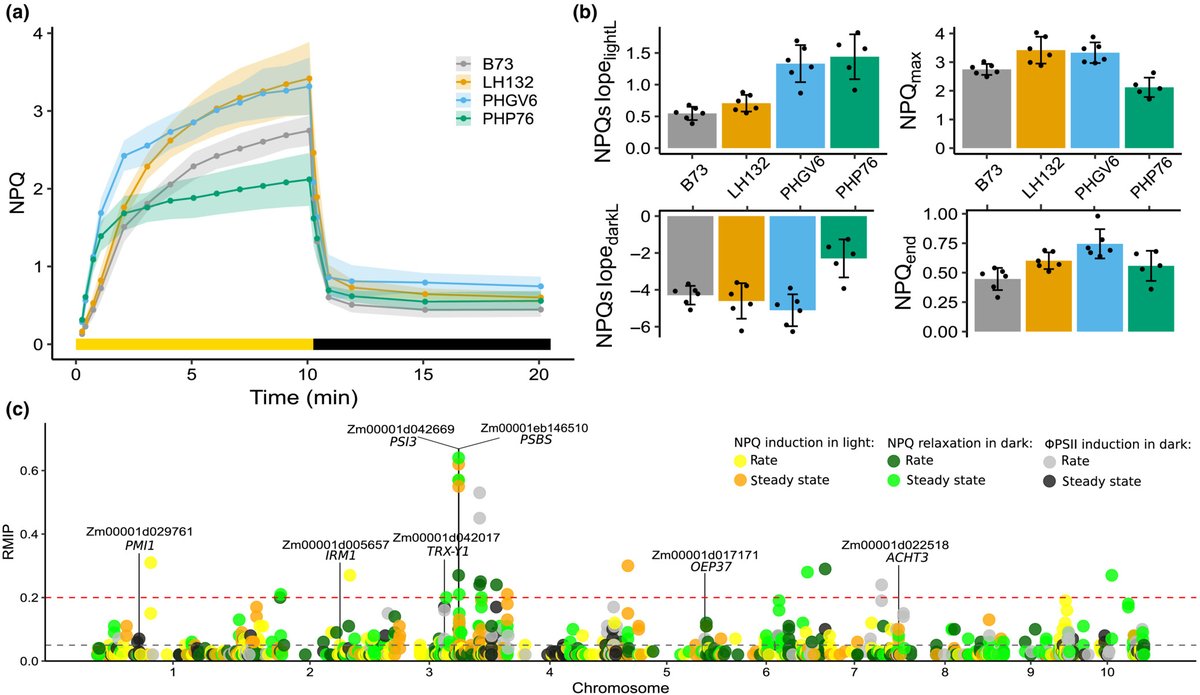
#EarlyViewAlert👁🗨Nonphotochemical quenching kinetics GWAS in sorghum identifies genes that may play conserved roles in maize and Arabidopsis thaliana photoprotection doi.org/10.1111/tpj.16… 🌽🌾 A work by Seema Sahay, @NikeeShrestha2, James Schnable et al University of Nebraska-Lincoln