Mario Suva
@mariosuva
Brain cancer biology. Single-cell genomics. Mass General Hospital. Broad Institute.
ID: 1151944815229190144
http://suvalab.mgh.harvard.edu/ 18-07-2019 20:00:20
890 Tweet
2,2K Followers
380 Following




12/ Before that, I would like to say thank wonderful co-first authors, @AvishaySptizer, Kevin Johnson and Luciano Garofano. I thank my great mentor Mario Suva and outstanding co-PIs Tirosh Lab, Roel Verhaak, Antonio Iavarone and Anna Lasorella.



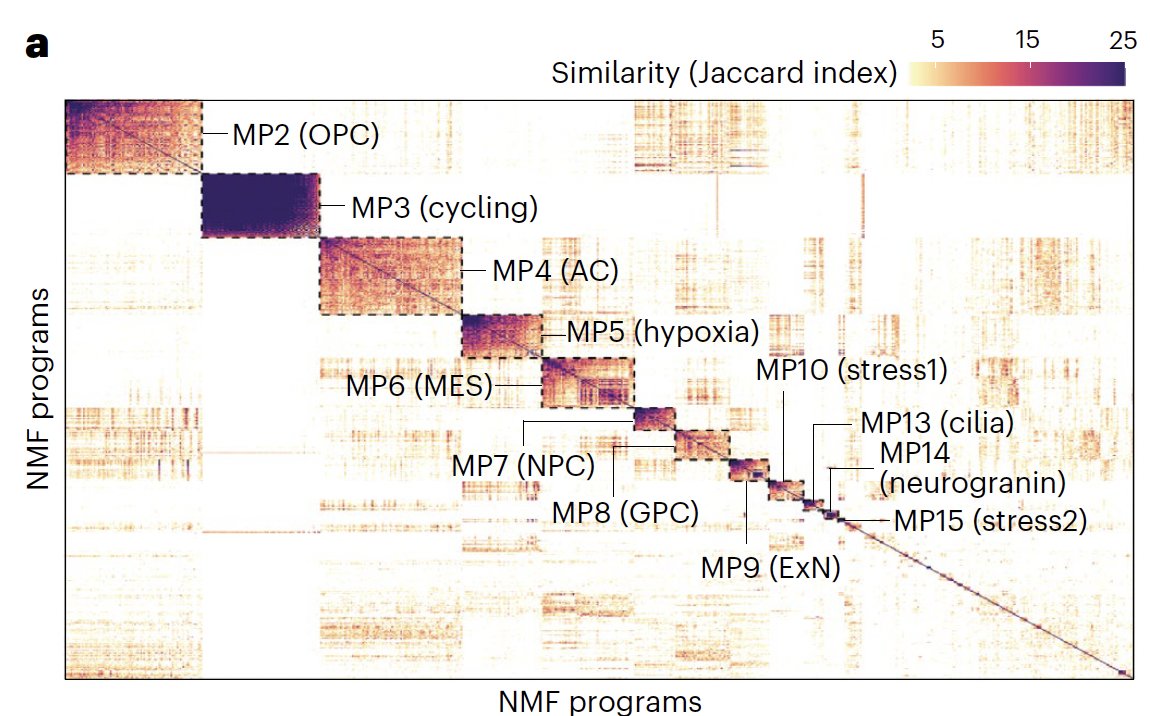
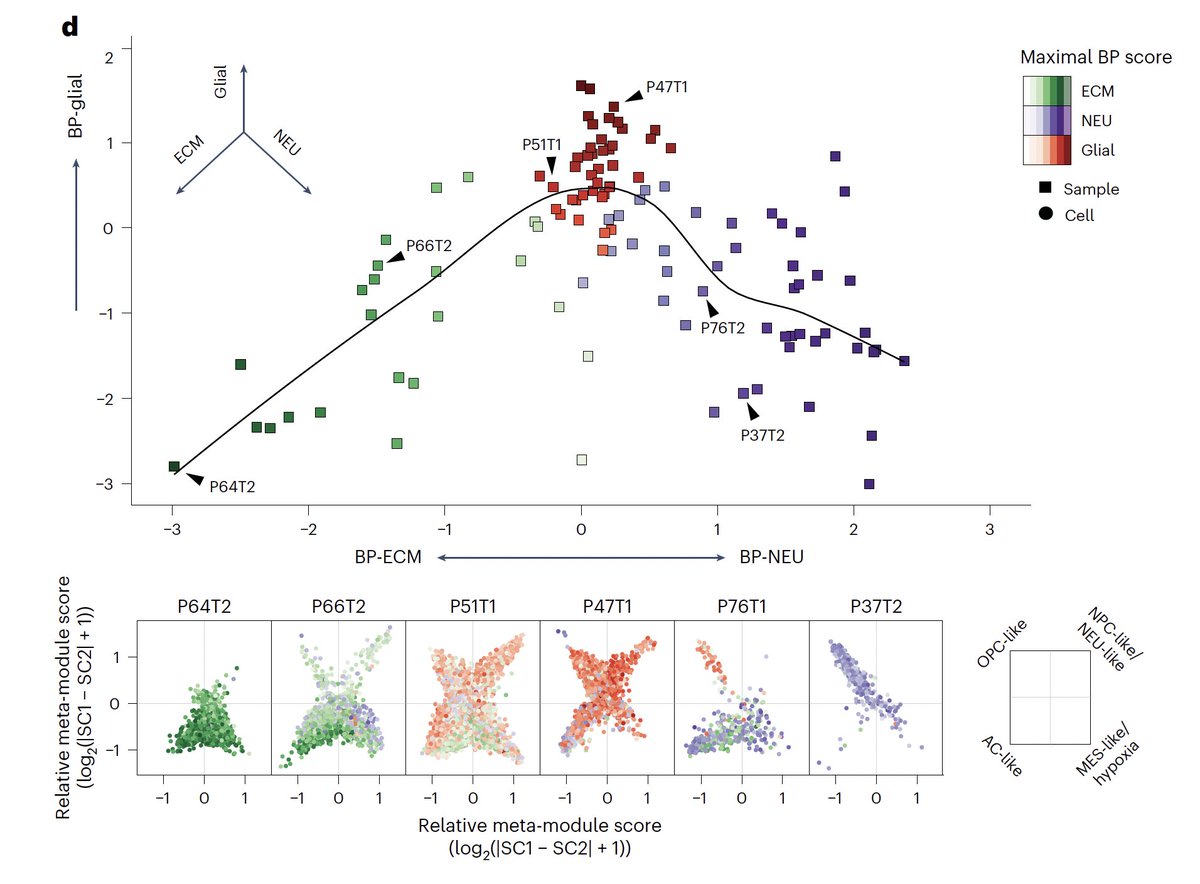
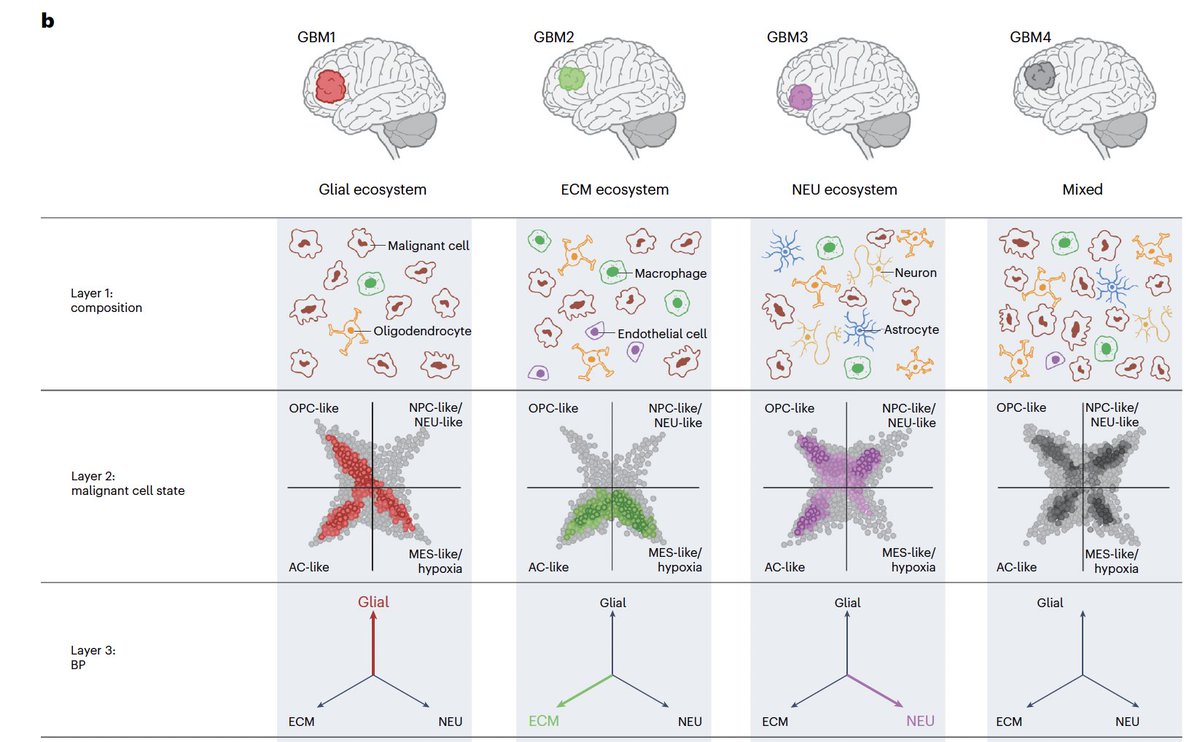
1/ Truly excited to share our new study that I had to privilege to co-lead during my PhD alongside great friends and collaborators Masashi Nomura, Kevin C. Johnson and Luciano Garofano, which was published Nature Genetics! nature.com/articles/s4158…











12/ Big thanks to the PIs that supervised the project Tirosh Lab, Mario Suva, Roel Verhaak, Antonio Iavarone and Anna Lasorella, all collaborators and institutions involved in the CARE consortium Weizmann Institute, MGH Pathology, Broad Institute, Yale School of Medicine and Miami University.

13/ Big thanks also to the institutions that provided tumor samples Duke University, MD Anderson Cancer Center, Tokyo University Hospital, Pitié-Salpêtrière Hospital, St. Michael's Hospital, Seoul National University and NORLUX and funders NIH, National Cancer Institute, Mark Foundation and Sontag Foundation.