
Mingze Dong
@mingze7316
PhD student @YaleCBB; Integrate Science B.S. @PKU1898
ID: 1453373401083568130
https://mingzedong.github.io 27-10-2021 14:50:21
20 Tweet
87 Followers
334 Following

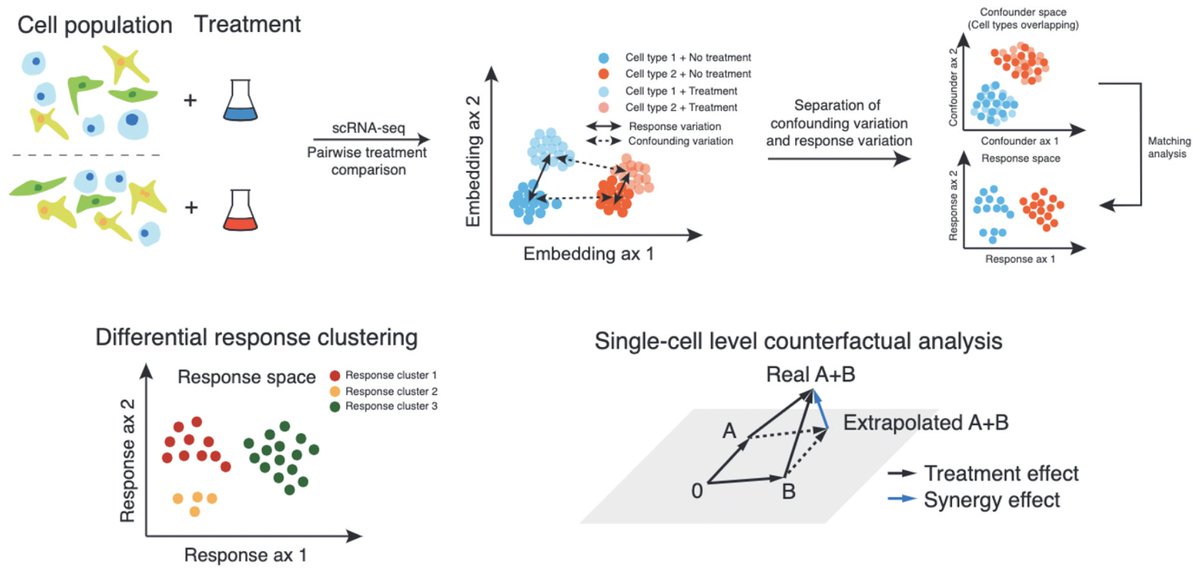
We are excited to present our latest project: CINEMA-OT – a causal inference framework for single-cell perturbation analysis! W/ Mingze Dong Rahul Dhodapkar Jeffrey Ishizuka Ellen Foxman biorxiv.org/content/10.110…

A very long overdue thread: happy to share preprint led by Sebastian Damrich from Fred Hamprecht's lab. *From t-SNE to UMAP with contrastive learning* arxiv.org/abs/2206.01816 I think we have finally understood the *real* difference between t-SNE and UMAP. It involves NCE! [1/n]
![Dmitry Kobak (@hippopedoid) on Twitter photo A very long overdue thread: happy to share preprint led by Sebastian Damrich from <a href="/FredHamprecht/">Fred Hamprecht</a>'s lab.
*From t-SNE to UMAP with contrastive learning*
arxiv.org/abs/2206.01816
I think we have finally understood the *real* difference between t-SNE and UMAP. It involves NCE! [1/n] A very long overdue thread: happy to share preprint led by Sebastian Damrich from <a href="/FredHamprecht/">Fred Hamprecht</a>'s lab.
*From t-SNE to UMAP with contrastive learning*
arxiv.org/abs/2206.01816
I think we have finally understood the *real* difference between t-SNE and UMAP. It involves NCE! [1/n]](https://pbs.twimg.com/media/Fi922YhXkAAZy7Y.jpg)

Spatial epigenome–transcriptome co-sequencing🤩🤩🤩Beyond excited to see it out today nature! Enjoyable collaboration between my lab @YaleBME @YaleSEAS Yale School of Medicine and GonçaloCasteloBranco of Karolinska Institutet. The most informative tool for spatial multi-omics!! nature.com/articles/s4158…




