Misha Kopylov
@mishakopylov3
SEMC @ NYSBC
ID: 1088134571508543488
23-01-2019 18:01:13
26 Tweet
127 Followers
52 Following



SEMC #NRAMM Chase Alex Wei Daija Misha Kopylov Edward T Eng with Peter Kahn (Engineering Arts), Jenny Hinshaw lab (NIH), Crina Nimigean lab (WCMC) & Seth Darst lab (Rockefeller). Using automation to solve macromolecular structure by #cryoEM nramm.nysbc.org








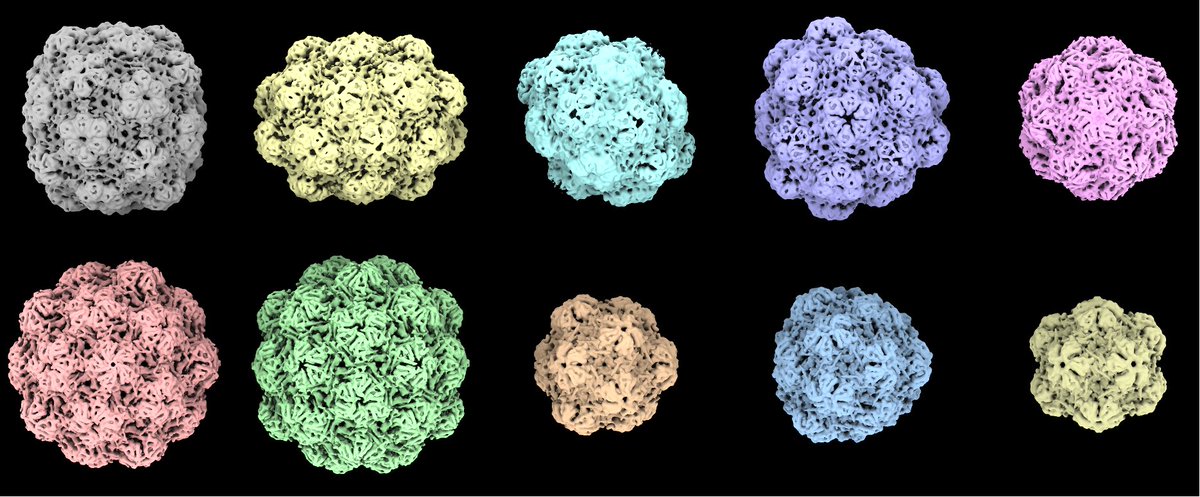
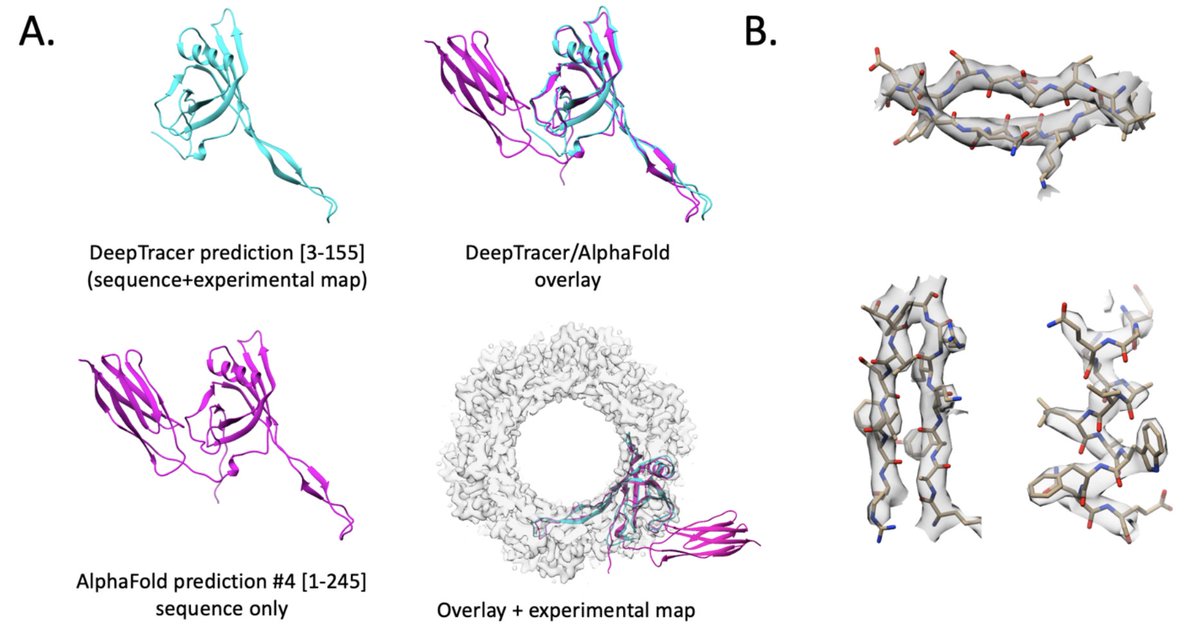
1 dataset – 10 structures! Check out the latest preprint by Sonia Bhattacharya from M.G. Finn’s group from Georgia Tech. Structural work was done by Matt Jenkins in collaboration with Carolina Hernandez and Misha Kopylov from NYSBC biorxiv.org/content/10.110…