Gonçalo C. Pereira
@mitopereira_gc
Project Scientist & Cell Assay Developer working in Drug Discovery @Astellas_ESM-UK . Tweets and opinions are my own.
ID: 247813782
https://www.linkedin.com/in/pereiragc/ 05-02-2011 16:37:00
530 Tweet
254 Followers
578 Following

I see so many new TPD agents reported that are supported by datasets which range massively in quality, so I’ve put together a simple guide for those in the field to help them identify the good agents from the rest using just 3 simple tests. See my blog. janusdrugdiscovery.com/240125-quick-g…

📣 We’re excited to announce our latest agreement with Astellas subsidiary, Nanna Therapeutics! Under the agreement, Nanna will access our DUB-focused #drugdiscovery platform to support the development of novel therapeutic candidates for multiple disease targets: rb.gy/dg71e4



Dr Tia Salter (she/her) and Will Allen's Bristol Biochemistry new paper: adaption of NanoLuc protein transport assay enables in vivo screening for inhibitors of bacterial secretion doi.org/10.1101/2024.0…… @bbsrc and Wellcome funded


Delighted to share the last study of the lab led by the fantastic postdoc Luis Carlos Tábara Rodríguez out in Cell !!! We show how MTFP1 controls mitochondrial fusion to regulate inner membrane quality control and maintain mtDNA levels. MRC MBU Cambridge University authors.elsevier.com/sd/article/S00…





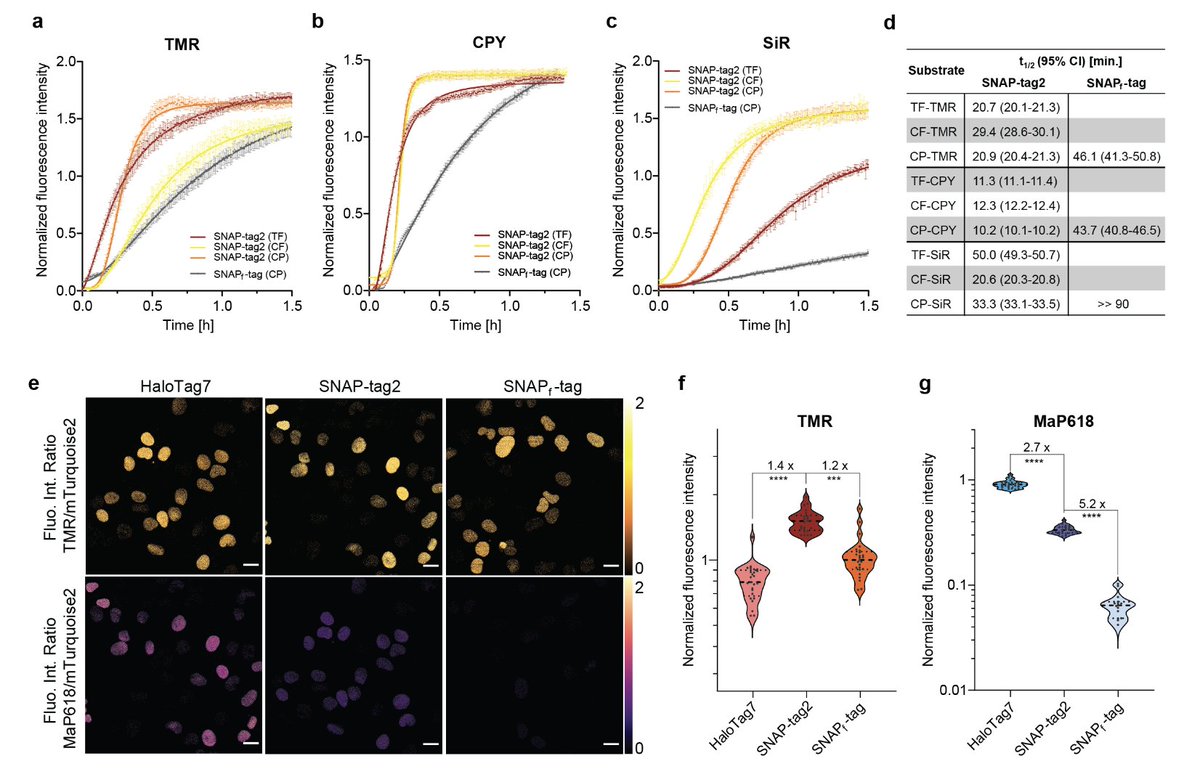
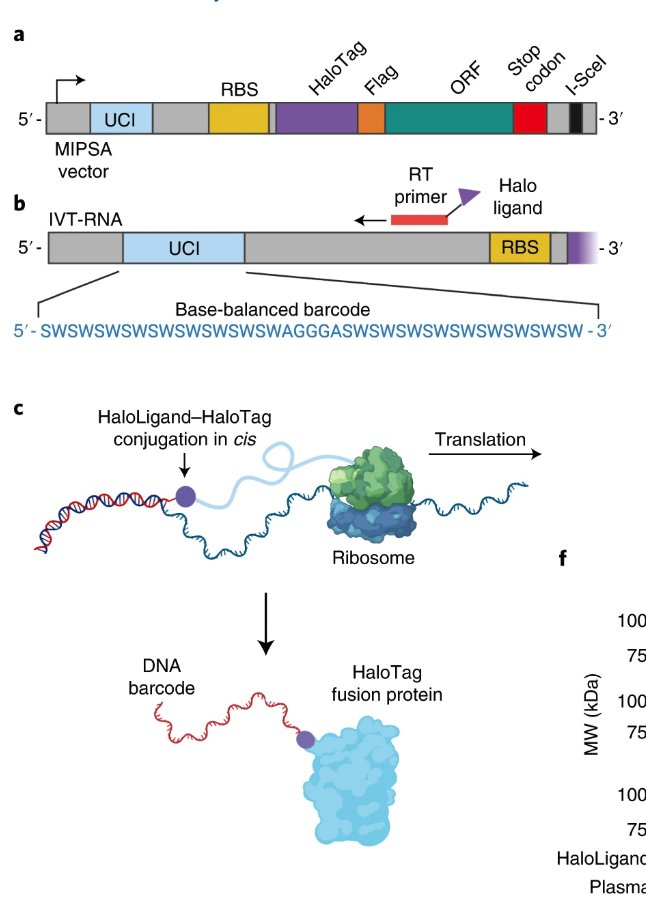
Want to degrade SNAP-tagged proteins? Take a look at our SNAP-#PROTACs. Great collaboration with Mariell Pettersson and team, Francesca Bottanelli lab, ACE from ZMB CRC 1430 Congrats and a big thank you to everyone who contributed! biorxiv.org/content/10.110…




Check out @JamesLorriman 's paper on the mitochondrial import/cleavage of PINK1, including lots of surprises! Super collaboration with Collinson Lab Robin Corey @robincorey.bsky.social & adam grieve (Bristol Biochemistry University of Bristol School of PPN University of Bristol) Wellcome PhD funding (James) biorxiv.org/content/10.110…