
Mohammed AlQuraishi
@moalquraishi
MLing biomolecules en route to structural systems biology. Asst Prof of Systems Biology and CS @Columbia. Prev. @Harvard SysBio; @Stanford Genetics, Stats.
ID: 925907616
http://aqlab.io 04-11-2012 18:50:33
1,1K Tweet
11,11K Followers
349 Following

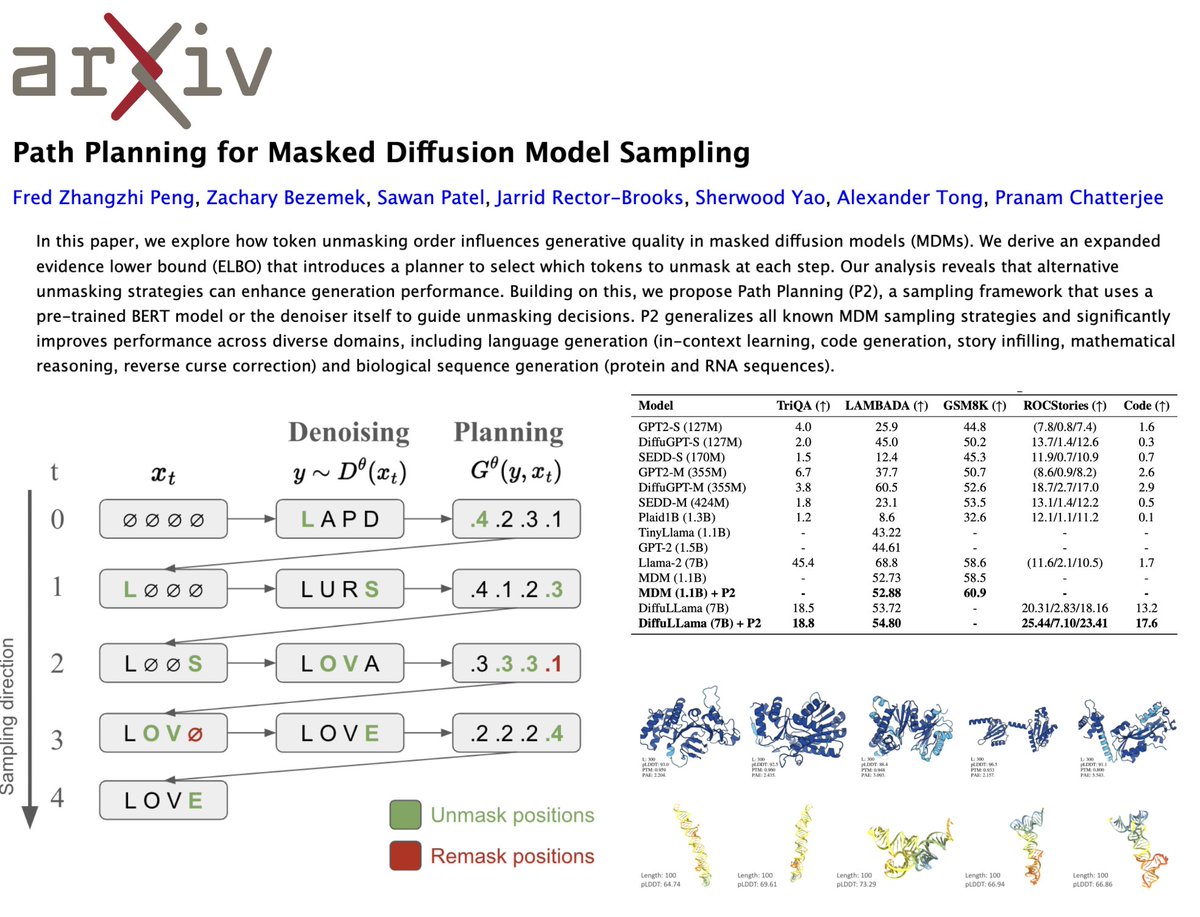
It's tough times for science. 🥺 But we have to keep innovating to fight another day, and today I'm so proud to share Fred Zhangzhi Peng's new, groundbreaking sampling algorithm for generative language models, Path Planning (P2). 🌟 📜: arxiv.org/abs/2502.03540 💻: In the appendix!!




Can confirm that Etowah Adams is awesome :-)

Alisia Fadini Minhuan Li ROCKET sounds like a game-changer for structural biology! Combining AlphaFold with crystallographic & cryoEM/ET data to refine models at scale is a huge step forward. Looking forward to seeing how this approach tackles the challenges of turning raw data into atomic models.



Excited to share our preprint “BoltzDesign1: Inverting All-Atom Structure Prediction Model for Generalized Biomolecular Binder Design” — a collaboration with Martin Pacesa, Zhidian Zhang , Bruno E. Correia, and Sergey Ovchinnikov. 🧬 Code will be released in a couple weeks



Proud that this work was accepted at ICML as a spotlight poster! Reminder that you can just do things, especially in interpretability. Etowah Adams and I did most of this work in colab notebooks that cost a few dollars a month.









