
OBO Foundry
@obofoundry
The Open Biological and Biomedical Ontology (OBO) Foundry
Community development of interoperable ontologies for the biological sciences
ID: 722459851037806592
http://obofoundry.org/ 19-04-2016 16:20:25
127 Tweet
374 Followers
86 Following

Our work on designing OBO Foundry ontology extensions for natural products won the Best Poster Award from the Bioontologies COSI at ISMB/ECCB 2021 and is now available - doi.org/10.5281/zenodo… NaPDI

We have developed the ontology for Avida digital evolution platform (gitlab.com/fortunalab/ont…), and it has been accepted to be part of the OBO Foundry (obofoundry.org/ontology/ontoa…). It's our fortunalab.org first contribution in the field of semantic databases.

We just added 63 new prefixes that cover a significant part of the missing prefixes used in OBO Foundry ontologies. Check out the huge PR here: github.com/biopragmatics/…

Thanks for your help shaping this #biodata primer, providing use cases & sharing w/ #datamanagement folks! ISO TDWG OBO Foundry GBIF @biodiversity.social/@gbif OBIS U.S. IOOS Office EDI DataONE Planet Microbe WoRMS ITIS.gov Data.gov NOAA NCEI


The @Bioregistry has an API that links entities to external resolvers and providers like in OBO Foundry identifiers.org EBISPOT OLS: 🧬 TP53 bioregistry.io/api/reference/… 🧪 Aspirin bioregistry.io/api/reference/… 🐕 Dog bioregistry.io/api/reference/… 🦠 COVID-19 bioregistry.io/api/reference/…


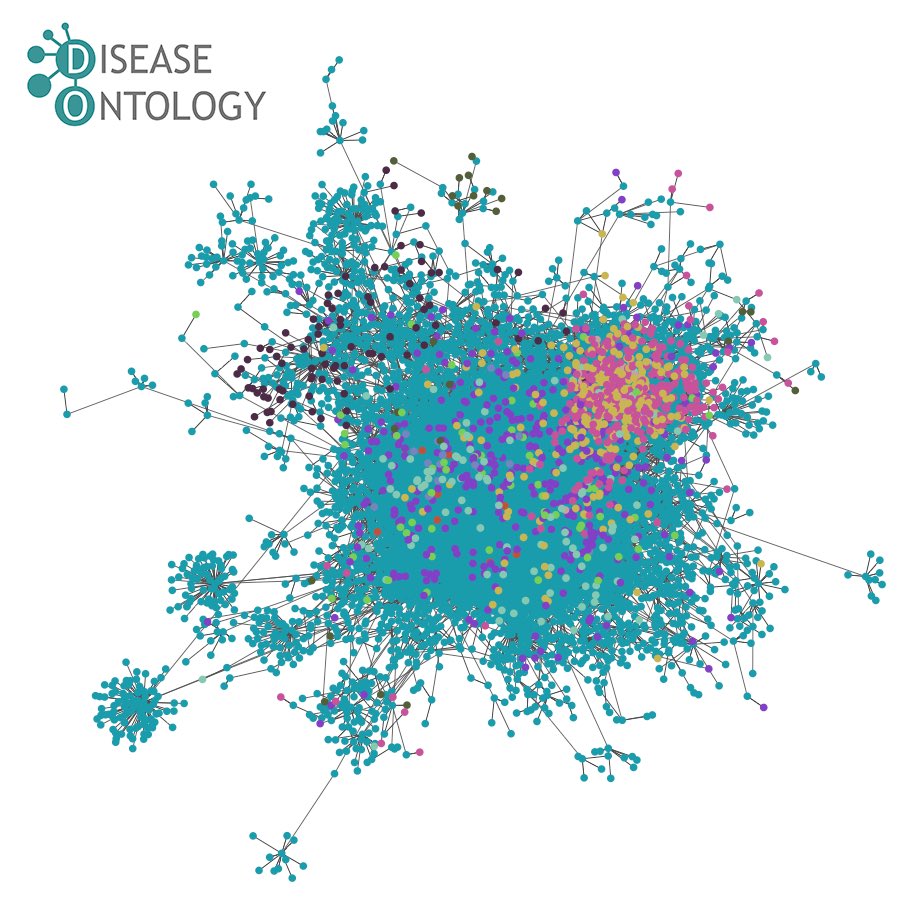
The Human Disease Ontology (DO) Knowledgebase has been #NIHfunded under a new National Human Genome Research Institute U24 $3.7M award to provide a comprehensive #disease feature similarity network for clinical differential diagnosis and biomedical data exploration. reporter.nih.gov/search/NKzIluA… #DiseaseResearch



Monarch Initiative HPO and #deepPhenotyping in our #exomiser tool improves #rareDisease #diagnostics! NIH ORIP National Human Genome Research Institute Rare Disease UK National Organization for Rare Disorders (NORD)

We have released version 0.3.0 of the #OntologyAccessKit #oaklib github.com/INCATools/onto…. Includes preliminary compatibility with cytoscape CX format and The NDEx Project, upgrade to the latest Bioregistry, and more support for working with the OBO Foundry #RelationOntology!

Here's our pre-print describing our GPT-3 based knowledge extraction tool SPIRES: arxiv.org/abs/2304.02711. Great work from J. Harry Caufield et al! SPIRES allows you to specify a knowledge schema (in LinkML - Linked (Open) Data Modeling Language) and then populate instances of that schema from unstructured text
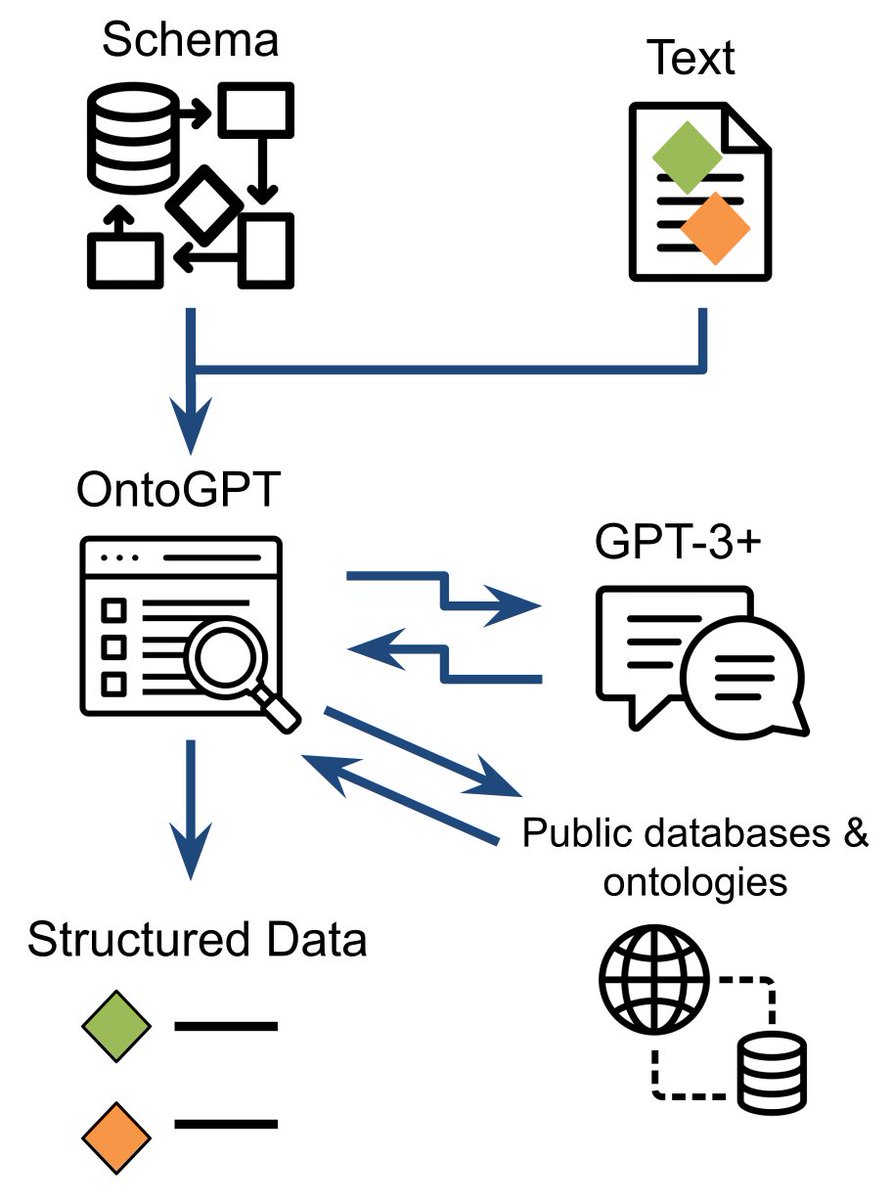

We want the results to use OBO Foundry class IDs from ontologies like HPO Gene Ontology @[email protected]. However, so far GPT-3 sucks as ontology term IDs (or rather it will hallucinate plausible sequences of digits that bear no relation to the actual term)...



Many thanks to our 2023 expert review panel and chair, BF Francis Ouellette, the team at GlobalBiodata, especially @RachelDrysdale4 and David Carr, and our supportive Member funding organisations.

SILVA was selected as Global Core Biodata Resource! See our acknowledgements here: arb-silva.de/news/view/2023… 🥳🎉 Congratulations to the other resources selected! Leibniz-Institut DSMZ LPSN - lpsn.dsmz.de Ribocon Max Planck Institute for Marine Microbiology


Congratualtions. UniProt has been a contributing member of the imexconsortium for 15 years and happy to be part of this valuable effort.

The Disease Ontology and DO-KB are thrilled to join the GCBR GlobalBiodata ontology OBO Foundry resources: Plant Ontology - Planteome Project Planteome project, Gene Ontology Gene Ontology @[email protected], ChEBI ChEBI as together we're dedicated to continue propelling global biomedical research forward!
