PCF-OUS Oslo
@pcfousoslo
Proteomics core facility Oslo, Immunology, Phosphoproteomics, Targeted proteomics
ID: 1180108025262198785
04-10-2019 13:11:39
41 Tweet
134 Followers
163 Following


Use of Proteomics and Phosphoproteomics demonstrate Sachin Singh GAK and PRKCD are positive regulators of PRKN-independent mitophagy biorxiv.org/content 10.1101/2020.11.05.369496v1

Planning to perform Proximity dependent biotinylation and wondering, who can help you to perform proteomics analysis . Please contact PCF-OUS Oslo and Sachin Singh. we have good expertise in wet-lab and data analysis.



Very good start of new year! Good collaboration between Jorunn Stamnæs and our proteomics core facilities PCF-OUS Oslo Sachin Singh NAPI Norway


We PCF-OUS Oslo are adding new BrukerMassSpec MS instrument to our PCF facility. Installation start today and we are looking forward to use it in coming weeks👏👏Sachin Singh NAPI Norway


Very productive day, installation process is much more faster than expected. Most difficult part was decide this amazing things a name. Finally, we named it Hilton II ( in-house name)NAPI Norway Sachin Singh Jrbrtsn


Read about three new BrukerMassSpec instruments just installed at the NAPI core facilities @unioslo_mn UiO - Det medisinske fakultet/Oslounivsykehus, and @UniNMBU. Exciting new investments that will benefit many projects. Thanks to Norges forskningsråd for funding. bit.ly/3jOzTKQ


It has been amazing week! PCF prepared more than 400 proteomics samples in week. We have developed high throughput sample preparations pipeline. If you want to investigate outcome of your drug or gene editing screen at protein level. We can help you with that. Sachin Singh


MaxDIA is in the spotlight of Nature Methods Many thanks to Dr. Arunima Singh for this kind overview doi.org/10.1038/s41592… #proteomics Max Planck Institute of Biochemistry (MPIB) MaxQuant Nature Biotechnology



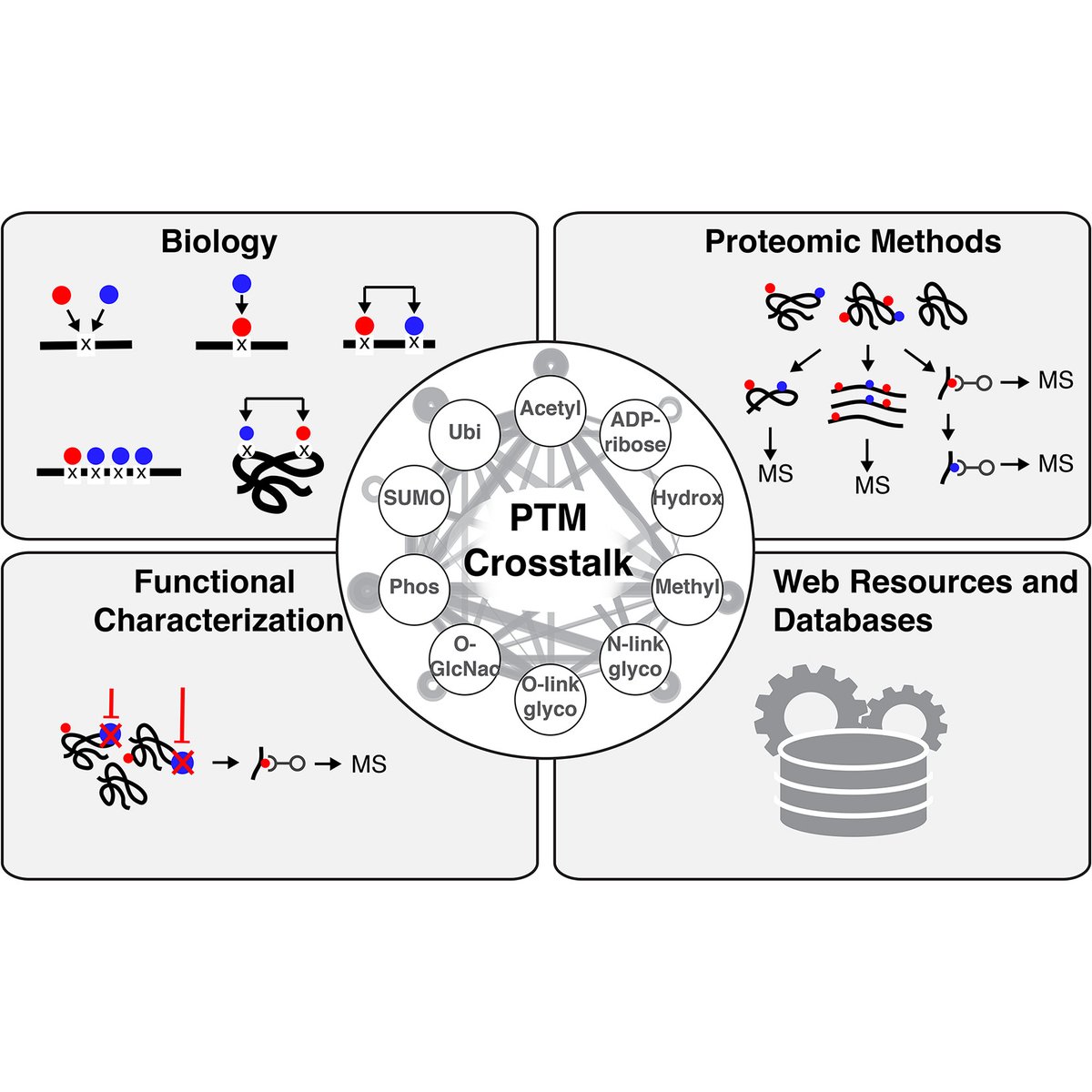
Leutert et al. provide an overview of the basic modes of #PTM crosstalk, the proteomic methods to elucidate PTM crosstalk, and approaches that can inform about the functional consequences of #PTMcrosstalk ― University of Washington Mario Leutert Judit Villen Sam Entwisle mcponline.org/article/S1535-…


Please read this nice article from Christopher Carroll and collaboration with PCF-OUS Oslo NAPI Norway