
Falk Ponath
@ponathf
ID: 1029270350616514560
https://www.helmholtz-hzi.de/en/research/research_topics/bacterial_and_viral_pathogens/rna_biology_o 14-08-2018 07:35:49
106 Tweet
171 Followers
388 Following

First paper from the Adams lab NICHD News is out Nature Communications! We comprehensively mapped RNA and characterized transcription termination in the Lyme pathogen B. burgdorferi — hundreds of truncated transcripts and potential regulators! nature.com/articles/s4146…
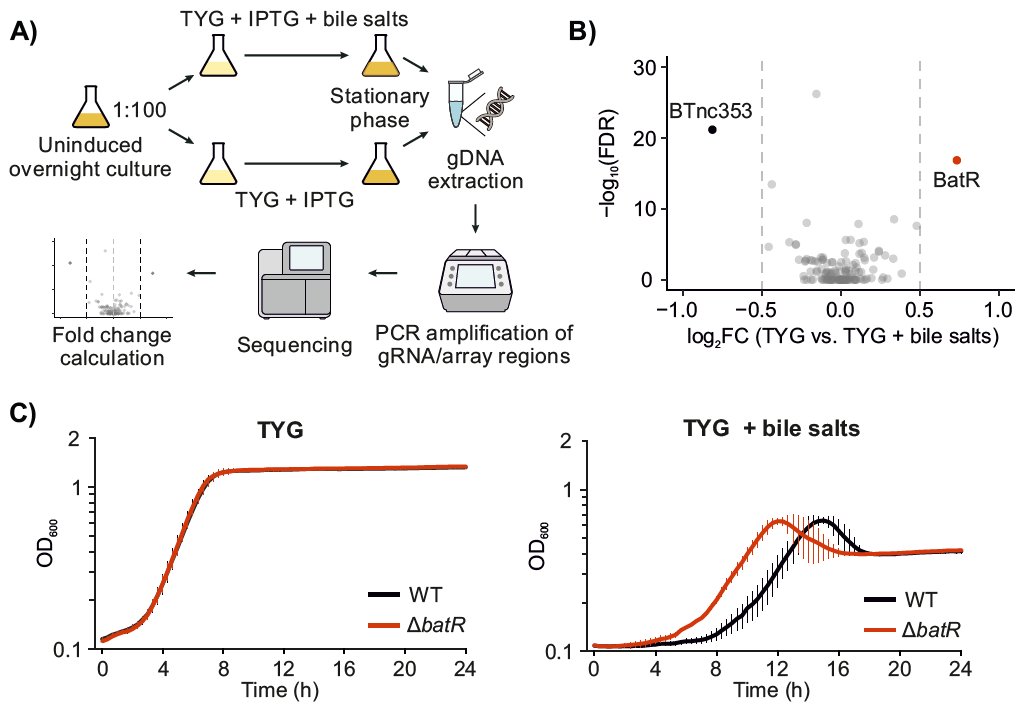
Our latest work, from @prezzag and in collaboration with Beisel Lab: we constructed a CRISPRi library of the intergenic sRNAs of B. theta and used it to identify modulators of growth under bile. Presenting BatR, the third sRNA we have characterized so far! biorxiv.org/content/10.110…

I ventured into the new world of RNA at Jörg Vogel with great support from Claudia Höbartner Lars Barquist Jakob Jung & the other co-authors! Preprint: A side-by-side comparison of peptide-delivered antisense antibiotics employing different nucleotide mimics biorxiv.org/content/10.110…

A conveyor belt of DCs: FLT3L regulates the spatiotemporal cascade of DC-development in LNs. Fascinating work from ka-mu-lab Milas Ugur out Immunity, great teamwork WüSI - Systems Immunology Würzburg Dominic Grün Emmanuel Saliba - open access cell.com/immunity/fullt…

Happy to share our latest work: Fusobacterium are naturally competent through Type IV pili. Great, concise study led by Blake Sanders and others not on twitter. We discovered it, now you go figure out what to do with it. sciencedirect.com/science/articl…
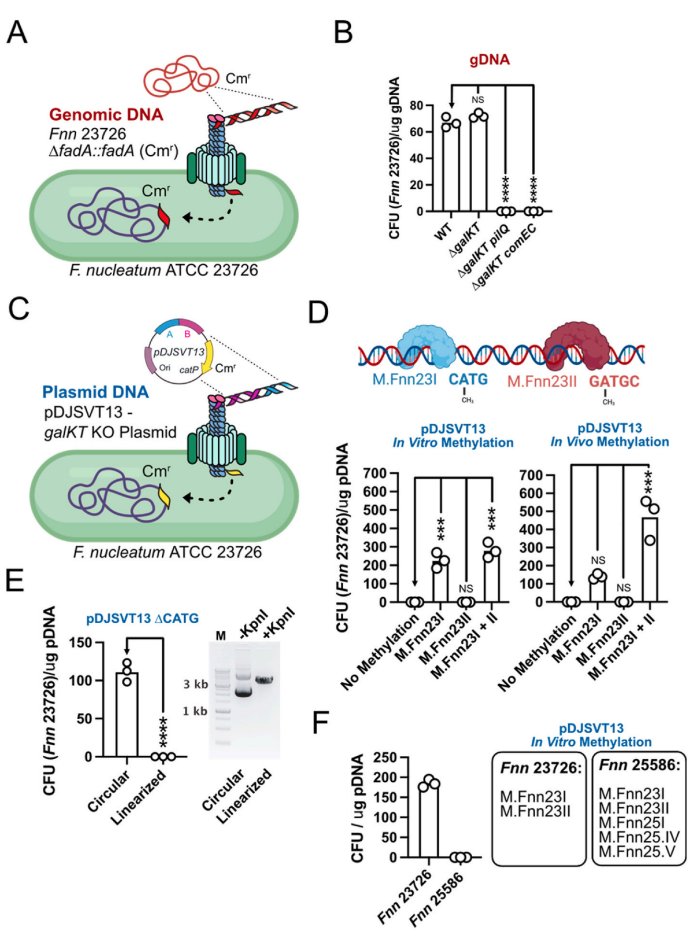

A new preprint of my PhD project Jörg Vogel working with TeamFuso Falk Ponath and Valentina Cosi, identifying two KH domain proteins-KhpA and KhpB as broad RBPs in Fusobacterium nucleatum.

Proud to present SHIFTR, a novel platform for region-specific RNA interactome discovery developed by fantastic PhD student @jensaydin 👨🔬 in my group, now published in Nucleic Acids Res: 1/8 academic.oup.com/nar/advance-ar…


Happy to share our latest work, spearheaded by @prezzag in collaboration with Beisel Lab Helmholtz Institute Würzburg (HIRI) and just out PNASNews. We set up a CRISPRi system in B. thetaiotaomicron and used it to screen sRNAs for their functions. [1/5] pnas.org/doi/10.1073/pn…

Another great collaboration with @Axel_R_Huber, @Cayetanopm, Joske Ubels, Hans Clevers, @BoxtelLab and many others. Random forest trained with #organoid #colibactin mutations => identification of pks-linked mutations in every 8th #colorectal tumor genome! authors.elsevier.com/sd/article/S15….

New research from Susan Bullman /CJohnstonLab, with superstar 1st author @mzepedar, out now in nature pinpoints “high-risk” subtype of Fusobacterium nucleatum that dominates the colorectal cancer (CRC) niche in human patient's Link : rdcu.be/dBUjH Tweet🧵below:

A new study from Bullman and CJohnstonLab Fred Hutch Cancer Center out today nature. We identify a 'high-risk' clade of F. nucleatum that dominates the human CRC niche. Huge congrats to the amazing first author @mzepedar and the incredible team. 🧵 from Chris 👇 nature.com/articles/s4158…

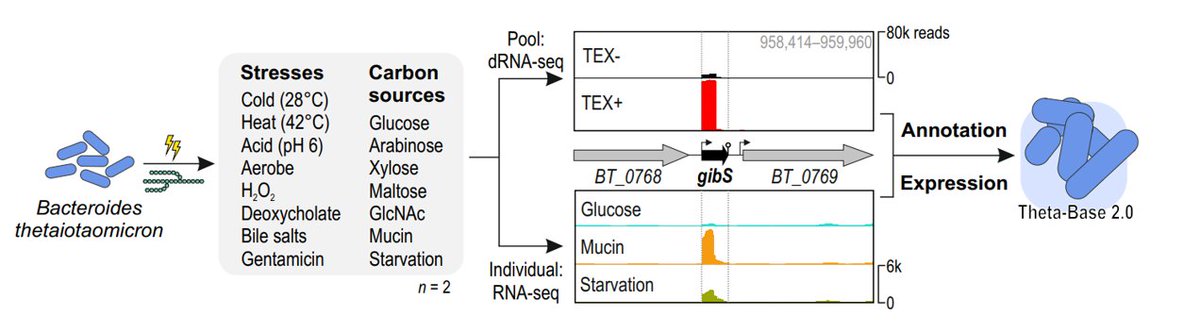
1/5: Molecular intricacies of gut bacteria! 🧬 Our study in Nature Microbiology , with colleagues from Helmholtz Institute Würzburg (HIRI), @Deutschbauer_Lab , & Lars Barquist groups, unveils B. thetaiotaomicron's transcriptional landscape across 15 gut conditions. nature.com/articles/s4156…


Check out the spectacular work led by Youssef El Mouali - giving insights into #sRNA regulation in the unexplored #Segatella/#Prevotella impacting metabolism and interactions with the host & commensals in the gut🦠

Small RNA w/big roles in colonization Youssef El Mouali StrowigLab characterize RNA landscape of Segatella copri. ID small RNA regulator essential for colonization & whose expression is regulated by metabolism of dietary components by S. copri cell.com/cell-host-micr…
