Eva Maria Steiner-Rebrova 🇺🇦🇦🇹🇸🇪
@rebrovaeva
🦠 bacteria, viruses and a host! on tip toes behind their powerful armoury and how we can re-program them for targeted therapy and as biologicals ;) @HalbergLab
ID: 1261999148087336962
17-05-2020 12:37:12
550 Tweet
175 Followers
251 Following




Our research group at Human Technopole , supported by European Research Council (ERC) aims to unravel the molecular mysteries of #thyroid hormone signalling. We seek for a passionate Postdoc to redefine paradigms in thyroid cell biology&disease 👉Apply now! lnkd.in/dbk_ePxH










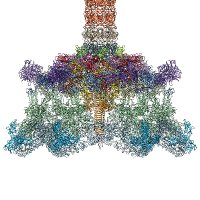
Exited to share our preprint on the structure of a Salmonella contractile injection system 🥳 Wonderful work by first author Rooshanie Ejaz Ejaz and Claudia Kielkopf, Freddie Martin, Marta Siborova, Eva Maria Steiner-Rebrova 🇺🇦🇦🇹🇸🇪, great collab with Marc Erhardt CPRatUCPH #cryoem shorturl.at/QHLpm

Excited to see our work published in Science Advances! We present RBPseg, a method for structural prediction of #phage receptor-binding proteins. We applied it to diverse tail fibers and validated the predicted structures using #cryoEM. Read here: science.org/doi/10.1126/sc…